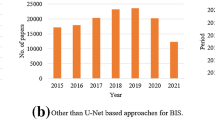
Abstract
Biomedical image segmentation is a mechanism that distinguishes the boundaries of various lesion regions within the image, be it 2D or 3D. In the current scenario, various soft and hard computing approaches have been used for medical image segmentation purposes. This article analyzes deep learning-based U-Net architecture for efficient segmentation by possible variations of its different important hyper-parameters that affect the U-Net performance. This article also investigates U-Net architecture’s performance to generate the segmented output over two diverse biomedical imaging datasets, such as brain MRI scans and serial section Transmission Electron Microscopy (ssTEM) dataset of the drosophila first instar larva VNC, hence making 48 possible combinations of execution of the U-Net model. The performance of the segmentation process has been evaluated using metrics like dice similarity coefficient and accuracy, while cross-entropy is considered the loss function. In doing so, an insight into the overall performance of U-Net on biomedical image segmentation problems has been obtained. The best results for accuracy and dice coefficient values out of all possible combinations made in this study come as 0.9881 and 0.9934, respectively. The main motive of this work is to provide an exhaustive and efficient analysis of the U-Net architecture for biomedical image segmentation and subsequent analysis.



















Similar content being viewed by others
References
Jena B, Nayak GK, Saxena S (2022) Convolutional neural network and its pretrained models for image classification and object detection: A survey. Concurrency and Computation: Concurr Comput Pract Exp. 34(6), e6767.
Jena B, Saxena S, Nayak GK, Saba L, Sharma N, Suri JS. Artificial intelligence-based hybrid deep learning models for image classification: the first narrative review. Comput Biol Med. 2021;137: 104803.
Jena B, Dash AK, Nayak GK, Mohapatra P, Saxena S. Image classification for binary classes using deep convolutional neural network: an experimental study. In: Trends of data science and applications. Berlin: Springer; 2021. p. 197–209.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. p. 234–41.
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, p. 3431–40.
Sinha P, Tuteja M, Saxena S. Medical image segmentation: hard and soft computing approaches. SN Appl Sci. 2020;2(2):1–8.
Jena B, Nayak GK, Saxena S. Comprehensive review of abdominal image segmentation using soft and hard computing approaches. In: 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA). IEEE; 2020. p. 1–5.
Saxena S, et al. Role of artificial intelligence in radiogenomics for cancers in the era of precision medicine. Cancers. 2022;14(12):2860.
FathiKazerooni A, et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci Rep. 2022;12(1):1–13.
Jena B, Nayak GK, Saxena S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Mach Vis Appl. 2022;33(1):1–16.
Kumari N, Saxena S. Review of brain tumor segmentation and classification. In: 2018 International Conference on Current Trends towards Converging Technologies (ICCTCT). IEEE; 2018. p. 1–6.
Saxena S, Garg A, Mohapatra P. Advanced approaches for medical image segmentation. In: Application of biomedical engineering in neuroscience. Berlin: Springer; 2019. p. 153–72.
Wu T, Manogaran AL, Beauchamp J, Waring GL. Drosophila vitelline membrane assembly: a critical role for an evolutionarily conserved cysteine in the “VM domain” of sV23. Dev Biol. 2010;347(2):360–8.
Gerhard S, Funke J, Martel J, Cardona A, Fetter R. Segmented anisotropic ssTEM dataset of neural tissue. figshare, 2013.
Cardona A, et al. An integrated micro-and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 2010;8(10): e1000502.
Wang C-W, Gosno EB, Li Y-S. Fully automatic and robust 3D registration of serial-section microscopic images. Sci Rep. 2015;5(1):1–14.
Gerhard S, Andrade I, Fetter RD, Cardona A, Schneider-Mizell CM. Conserved neural circuit structure across Drosophila larval development revealed by comparative connectomics. Elife. 2017;6: e29089.
Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35(5):1240–51.
Chang J, Zhang X, Ye M, Huang D, Wang P, Yao C. Brain tumor segmentation based on 3D Unet with multi-class focal loss. In: 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI). IEEE; 2018. p. 1–5.
Skourt BA, El Hassani A, Majda A. Lung CT image segmentation using deep neural networks. Procedia Comput Sci. 2018;127:109–13.
Das S, Nayak GK, Saxena S, Satpathy SC (2021) Effect of learning parameters on the performance of U-Net Model in segmentation of Brain tumor. Multimed Tools Appl. 1–19.
Das S, Bose S, Nayak G K, Satapathy SC, Saxena S (2021) Brain tumor segmentation and overall survival period prediction in glioblastoma multiforme using radiomic features. Concurrency and Computation: Concurr Comput Pract Exp, e6501.
Zhou X-Y, Yang G-Z. Normalization in training U-Net for 2-D biomedical semantic segmentation. IEEE Robot Autom Lett. 2019;4(2):1792–9.
Baheti B, Innani S, Gajre S, Talbar S. Eff-unet: a novel architecture for semantic segmentation in unstructured environment. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, 2020, p. 358–59.
Kolařík M, Burget R, Uher V, Dutta M.K. 3D dense-U-Net for MRI brain tissue segmentation. In: 2018 41st International Conference on Telecommunications and Signal Processing (TSP). IEEE; 2018. p. 1–4.
Falk T, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70.
Alom MZ, et al. A state-of-the-art survey on deep learning theory and architectures. Electronics. 2019;8(3):292.
Zou KH, et al. Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Acad Radiol. 2004;11(2):178–89.
Khadangi A, Boudier T, Rajagopal V. EM-net: deep learning for electron microscopy image segmentation. In: 2020 25th International Conference on Pattern Recognition (ICPR). IEEE; 2021. p. 31–38.
Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151(1):41–60.
Menze BH, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2014;34(10):1993–2024.
Bakas S, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4(1):1–13.
Bakas S, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. 2018. arXiv preprint http://arxiv.org/abs/02629.
Alqazzaz S, Sun X, Yang X, Nokes L. Automated brain tumor segmentation on multi-modal MR image using SegNet. Comput Vis Media. 2019;5(2):209–19.
Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–20.
Araujo FH, et al. Deep learning for cell image segmentation and ranking. Comput Med Imaging Graph. 2019;72:13–21.
Shibuya E, Hotta K. Feedback U-Net for cell image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, 2020, p. 974–5.
Funding
The authors declare that they are not receiving funds for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jena, B., Nayak, G.K., Paul, S. et al. An Exhaustive Analytical Study of U-Net Architecture on Two Diverse Biomedical Imaging Datasets of Electron Microscopy Drosophila ssTEM and Brain MRI BraTS-2021 for Segmentation. SN COMPUT. SCI. 3, 418 (2022). https://doi.org/10.1007/s42979-022-01347-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42979-022-01347-y