Abstract
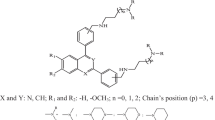
Free energy force field (FEFF) 3D-QSAR analysis was used to construct ligand-receptor binding models for a set of 18 structurally diverse antifolates including pyrimethamine, cycloguanil, methotrexate, aminopterin and trimethoprim, and 13 pyrrolo[2,3-d]pyrimidines. The molecular target (`receptor') used was a 3D-homology model of a specific mutant type of Plasmodium falciparum (Pf) dihydrofolate reductase (DHFR). The dependent variable of the 3D-QSAR models is the IC50 inhibition constant for the specific mutant type of PfDHFR. The independent variables of the 3D-QSAR models (the descriptors) are scaled energy terms of a modified first-generation AMBER force field combined with a hydration shell aqueous solvation model and a collection of 2D-QSAR descriptors often used in QSAR studies. Multiple temperature molecular dynamics simulation (MDS) and the genetic function approximation (GFA) were employed using partial least square (PLS) and multidimensional linear regressions as the fitting functions to develop FEFF 3D-QSAR models for the binding process. The significant FEFF energy terms in the best 3D-QSAR models include energy contributions of the direct ligand-receptor interaction. Some changes in conformational energy terms of the ligand due to binding to the enzyme are also found to be important descriptors. The FEFF 3D-QSAR models indicate some structural features perhaps relevant to the mechanism of resistance of the PfDHFR to current antimalarials. The FEFF 3D-QSAR models are also compared to receptor-independent (RI) 4D-QSAR models developed in an earlier study and subsequently refined using recently developed generalized alignment rules.
Similar content being viewed by others
References
The World Health Organization Report; Who Publications: Geneva, 1997.
Tokarski, J.S. and Hopfinger, A.J., J. Chem. Inf. Comput. Sci., 37 (1997) 779.
Tokarski, J.S. and Hopfinger, A.J., J. Chem. Inf. Comput. Sci., 37 (1997) 792.
Hopfinger, A.J., In Conformational Properties of Macromolecules, Academic Press, New York, NY, 1973, p. 71.
Koehler, M.G. and Hopfinger, A.J., Polymer, 30 (1989) 116.
Weiner, S.J., Kollman, P.A. and Nguyen, D.T.A., J. Comput. Chem., 7 (1986) 230.
Hopfinger, A.J. and Pearlstein, R.A., J. Comput. Chem., 5 (1984) 486.
Allinger, N.L., J. Am. Chem. Soc., 99 (1977) 8127.
Brobey, R.K.B., Iwakura, M., Itoh, F., Aso, K. and Horii, T., Parasitol. Int., 47 (1998) 69.
Blakley, R.L. In Blakley R.L. and Benkovic, S.J. (Eds), Folates and Pterins, Vol. 1, John Wiley & Sons, New York, NY, 1984, p. 191.
Kraut, J. and Matthews, D.A., In Jurnak F. and McPherson, A. (Eds), Biological Macromolecules and Assemblies, Vol. 3, John Wiley & Sons, New York, NY, 1987, p. 1.
Miller, G.P. and Benkovic, S.J., J. Chem. Biol., 5 (1998) R105.
Bzik, D.J., Li, W.-B., Horii, T. and Inserburg, J., Proc. Natl. Acad. Sci. USA., 84 (1987) 8360.
Burkhard, R. and Sander, C., Annu. Rev. Biophys. Biomol. Struct., 25 (1996) 113.
Šali, A., Curr. Opin. Biotechnol., 6 (1995) 437.
Johnson, M.S., Srinivasan, N., Sowdhamini, R. and Blundell, T.L., Crit. Rev. Biochem. Mol. Biol., 29 (1994) 1.
Davies II, J.F., Delcamp, T.J., Prendergast, N.J., Ashford, V.A., Freisheim, J.H. and Kraut, J., Biochemistry, 29 (1990) 9467.
Matthews, D.A., Bolin, J.T., Burridge, J.M., Filman, D.J., Volz, K.W. and Kraut, J., J. Biol. Chem., 260 (1985) 392.
Volz, K.W., Matthews, D.A., Alden, R.A., Freer, S.T., Hansch, C., Kaufman, B.T., Kraut, J., J. Biol. Chem., 257 (1982) 2528.
Bystroff, C., Oatley, S.J. and Kraut, J., Biochemistry, 29 (1990) 3263.
Sawaya, M.R. and Kraut, J., Biochemistry, 36 (1997) 586.
Bernstein, F.C., Koetzle, T.F., Williams, G.J.B., Meyer, Jr., E.F., Brice, M.D., Rodgers, J.R., Kennard, O., Schimanouchi, T. and Tasumi, M., J. Mol. Biol., 112 (1977) 535.
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N. and Bourne, P.E., Nucl. Acids Res., 28 (2000) 235. (http://www.rcsb.org/pdb/)
Laskowski, R.A., McArthur, M.W., Moss, D.S. and Thornton, J.M., J. Appl. Cryst., 26 (1993) 283.
Ramachandran, G.N. and Sassiekharan, V., Adv. Prot. Chem., 28 (1968) 283.
HyperChem Program Release 5.01 for Windows, Hypercube, Inc.; 1996.
Santos-Filho, O.A. and Hopfinger, A.J., J. Comput. Aid. Mol. Des., 15 (2001) 1.
Sano, G.-I., Morimatsu, K. and Horii, T., Mol. Biochem. Parasitol., 63 (1994) 265.
Rogers, D. and Hopfinger, A.J., J. Chem. Inf. Comput. Sci., 34 (1994) 854.
Rogers, D. WOLF Reference Manual Version 5.5, Molecular Simulation Inc., 1994.
Glen, W.G., Dunn, W.J., III and Scott, D.R., Tetrahedron Comput. Methods, 2 (1989) 349.
Rogers, D., In The Proceedings of the Fourth International Conference on Genetic Algorithms, San Diego, 1991, p. 38.
Rogers, D., personal communication, 1996.
Rogers, D., In Proceedings of the Seventh International Conference on Genetic Algorithms, East Lansing, MI, Morgan-Kaufmann, San Francisco, CA, 1997.
Doherty, D.C.; MOLSIM-User Guide; The Chem21 Group; 1780 Wilson Dr., Lake Forest, IL 60045, 1994.
Berendsen, H.J.C., Postman, J.P.M., van Gunsteren, W.F.; Di Nola, A. and Haak, J.R., J. Chem. Phys., 81 (1984) 3684.
Cerius 2 version. 3.0. Molecular Simulations Inc., 9685 Scranton Road, San Diego, CA 92121-3752, USA.
Albuquerque, M.G., Hopfinger, A.J. Barreiro, E.J. and de Alencastro, R.B., J. Chem. Inf. Comput. Sci., 38 (1998) 925.
Kier, L.B. and Hall, L.H., J. Chem. Inf. Comput. Sci., 40 (2000) 792.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Santos-Filho, O.A., Mishra, R.K. & Hopfinger, A. Free energy force field (FEFF) 3D-QSAR analysis of a set of Plasmodium falciparum dihydrofolate reductase inhibitors. J Comput Aided Mol Des 15, 787–810 (2001). https://doi.org/10.1023/A:1013199108020
Issue Date:
DOI: https://doi.org/10.1023/A:1013199108020