Abstract
Hypertension is a major risk factor for cardiovascular disease (CVD), yet blood pressure is measured intermittently and under suboptimal conditions. We developed a deep learning model to identify hypertension and stratify risk of CVD using 12-lead electrocardiogram waveforms. HTN-AI was trained to detect hypertension using 752,415 electrocardiograms from 103,405 adults at Massachusetts General Hospital. We externally validated HTN-AI and demonstrated associations between HTN-AI risk and incident CVD in 56,760 adults at Brigham and Women’s Hospital. HTN-AI accurately discriminated hypertension (internal and external validation AUROC 0.803 and 0.771, respectively). In Fine-Gray regression analyses model-predicted probability of hypertension was associated with mortality (hazard ratio per standard deviation: 1.47 [1.36-1.60], p < 0.001), HF (2.26 [1.90-2.69], p < 0.001), MI (1.87 [1.69-2.07], p < 0.001), stroke (1.30 [1.18-1.44], p < 0.001), and aortic dissection or rupture (1.69 [1.22-2.35], p < 0.001) after adjustment for demographics and risk factors. HTN-AI may facilitate diagnosis of hypertension and serve as a digital biomarker of hypertension-associated CVD.
Similar content being viewed by others
Introduction
Hypertension affects over 1 billion individuals worldwide1 and is a major modifiable risk factor for cardiovascular disease (CVD)2,3,4,5,6,7. Although measurement of blood pressure (BP) is considered straightforward, many patient- and measurement-related factors can influence BP measurements, contributing to high inter-visit variability in measured BP1,8,9,10. Society guidelines for accurate BP measurement are extensive and include requiring patients to rest for 5 minutes, ensuring the patient has emptied their bladder, avoiding exercise and caffeine prior to measurement, and averaging BP readings over multiple visits, among other recommendations1,8,9. However, these recommendations can be difficult to rigorously implement in busy clinical settings11.
Despite the high variability of BP measurement, clinicians typically diagnose and manage hypertension using office BP readings. In doing so, they must use clinical judgment to estimate a patient’s ambulatory blood pressure from sparse measurements taken in settings that are unrepresentative of the patient’s daily life. A further complication is that an estimated 10–15% of patients have masked hypertension, in which office BP is lower than ambulatory BP, with reported prevalence as high as 50% in African Americans1,12. While guidelines recommend 24-hour ambulatory BP monitoring to evaluate masked hypertension1,8,9, ambulatory BP monitoring is used infrequently13 and requires clinicians to suspect ambulatory hypertension justify the test. There is therefore a need for a convenient biomarker for ambulatory hypertension that integrates the effects of highly dynamic BPs over time, just as the hemoglobin A1C does for blood glucose.
One modality that provides information about ambulatory hypertension is the 12-lead electrocardiogram (ECG). Chronic hypertension is well-known to cause changes in cardiac structure and conduction that are reflected in ECG features such as increased QRS voltages, prolonged QT interval, prolonged P wave duration and PR interval, and abnormal repolarization14. However, outside of extreme features such as high voltages meeting criteria for left ventricular hypertrophy, the ECG features of hypertension are subtle and difficult to use in routine clinical practice.
In this study we aimed to train and validate a deep learning model, named HTN-AI, that identifies signals within the 12-lead ECG waveform associated with changes in cardiac structure and conduction caused by hypertension. We demonstrate 2 applications of HTN-AI. First, we show that HTN-AI can be used to identify hypertension using only the 12-lead ECG waveform as input. Second, because the adverse effects of hypertension are mediated in part by hypertension’s effect on the myocardium which may manifest in the ECG, we show that HTN-AI can serve as digital biomarker15 of hypertension-associated cardiovascular risk which can be used to stratify risk of CVD including mortality, heart failure (HF), myocardial infarction (MI), stroke, and aortic dissection or rupture.
Results
Population characteristics
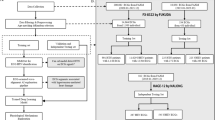
Figure 1 depicts an overview of the study. The Massachusetts General Hospital (MGH) sample comprised 121,720 patients which were divided into training, development, and internal validation samples (Table 1). There were 752,415 ECGs in the training and development samples, with median 3 (1–6) ECGs per patient and a median time difference between ECG and start of follow up of −121 (−561 to 409) days. Mean age in the MGH sample was 57.3 (16.8) years and 76,499 (62.8%) patients had known hypertension. Among those with office BP measurements (n = 36,718 [30.2%] missing systolic BP [SBP]; n = 36,725 [30.2%] missing diastolic BP [DBP]), 18,835 (22.2%) had SBP ≥ 140 mmHg, and 8,680 (10.2%) had DBP ≥ 90 mmHg. Prevalence of baseline cardiovascular comorbidities is shown in Table 1, and disease definitions are provided in Supplementary Table 1.
The HTN-AI model is a 3-dimensional convolutional neural network that was trained to identify hypertension using only 12-lead ECG waveforms in longitudinal primary care and cardiology patients at Massachusetts General Hospital. HTN-AI was validated in a sample of primary care patients from Brigham and Women’s Hospital. We also tested associations between the HTN-AI score (i.e. predicted probability of hypertension) and incident outcomes including all-cause mortality, heart failure, myocardial infarction, stroke, and aortic dissection and rupture in the Brigham and Women’s Hospital sample. BWH Brigham and Women’s Hospital, C3PO Community Care Cohort Project, ECG electrocardiogram, EWOC Enterprise Warehouse of Cardiology, MGH Massachusetts General Hospital.
The Brigham and Women’s Hospital (BWH) external validation and outcomes sample included 56,760 patients with mean age 55.1 (16.2) years. The prevalence of hypertension (31,014 [54.6%]) was lower, though the rates of elevated SBP (12,494 [23.4%]; n = 3339 [5.9%] missing) and DBP (6,240 [11.7%]; n = 3340 [5.9%] missing) were similar. Only the closest ECG before the start of follow up was used during external validation, with a median time difference of −533 (−723 to −279) days.
A digital biomarker for hypertension
We validated the use of the “HTN-AI score” (i.e. model predicted probability of hypertension, range 0–1) as a digital biomarker for hypertension diagnosis in 3 ways: (1) by validating model performance for identification of prevalent hypertension, (2) by demonstrating associations between the HTN-AI score and short-term incidence of hypertension diagnosis, and (3) by testing associations between the HTN-AI score and mean 24-hour ambulatory blood pressure.
First, we examined model performance in identifying prevalent hypertension using only the 12-lead ECG waveform. The HTN-AI score discriminated patients with baseline hypertension or elevated BP with an AUROC of 0.803 (0.796–0.810) in the MGH internal validation sample and 0.771 (0.767–0.775) in the BWH validation sample (Supplementary Fig. 2). Average precision was 0.865 (0.858–0.872; prevalence 0.655) in the MGH internal validation sample and 0.811 (0.806–0.816; prevalence 0.597) in the BWH external validation sample. At a HTN-AI score threshold of 0.85, which was defined to achieve 90% specificity in the model development sample to simulate an opportunistic screening use case, HTN-AI achieved internal validation sensitivity of 0.412 (specificity 0.908) and external validation sensitivity of 0.329 (specificity 0.917). Other performance metrics and model sensitivity analyzes are shown in Supplementary Table 2. Model performance metrics for model outputs other than the HTN-AI score is shown in Supplementary Table 3.
Second, we tested associations between the HTN-AI score and short-term incidence of new hypertension diagnosis as a proxy for the ability of the HTN-AI score to detect undiagnosed hypertension. Among 2282 patients in the high-risk HTN-AI group without baseline hypertension (i.e. HTN-AI “false positives”) there were 1,261 incident hypertension diagnoses (estimated 1-year cumulative incidence of 17.3% [15.7–18.9]) compared to 7351 diagnoses among 23,043 patients in the HTN-AI low-risk group (i.e. “true negatives”; estimated 1-year cumulative incidence of 7.1% [6.8–7.4]; p < 0.001; Supplementary Table 4), suggesting that the HTN-AI score has prognostic value for detection of hypertension among patients with suspected normal BP.
Third, we examined the relationship between HTN-AI risk and gold-standard measurement of mean 24-hour ambulatory BP. We identified 243 patients in the BWH sample who had both 24-hour ambulatory BP monitoring and a 12-lead ECG performed <1 year apart. Within this sample, mean systolic BP was 130.6 ± 15.4 mmHg and mean diastolic BP was 73.9 ± 10.7 mmHg. A total of 177/243 (72.8%) patients had elevated 24-hour average BP (i.e. average systolic BP ≥ 125 mmHg or average diastolic BP ≥ 75 mm Hg). High HTN-AI risk was associated with an increase in mean 24-hour systolic blood pressure of 11.0 (6.6–15.4) mmHg as compared to low HTN-AI risk, after adjustment for age, sex, and antihypertensive medication use. High HTN-AI risk was also associated with an odds ratio of 3.03 (1.47–6.64) for elevated 24-hour ambulatory BP after adjustment for age, sex, and antihypertensive medication use (p = 0.004).
Risk stratification of hypertension-associated cardiovascular disease
We explored whether HTN-AI can be used as a digital biomarker of hypertension-associated cardiovascular risk by investigating associations between the HTN-AI score and five sequelae of hypertension: mortality, HF, MI, stroke, and aortic dissection or rupture. HTN-AI risk significantly stratified incidence of all examined outcomes (Fig. 2 and Supplementary Fig. 3). For example, among 12,945 patients with high HTN-AI risk, 4181 died over median follow up of 5.6 years (estimated 10-year cumulative incidence 21.0% [20.2–21.7]). In comparison, among 43,225 patients at low HTN-AI risk, 4125 patients died over median follow up of 7.8 years (10-year cumulative incidence 5.4% [5.2–5.6]; p < 0.001). Event rates for other outcomes are shown in Supplementary Table 5.
In the Brigham and Women’s Hospital external validation sample, we assessed cumulative incidence of cardiovascular outcomes including (a) all-cause mortality, b heart failure, c myocardial infarction, and d ischemic stroke, stratified by HTN-AI risk. Cumulative incidence of aortic dissection or rupture is shown in Supplementary Fig. 3. HTN-AI risk was able to significantly stratify cumulative incidence of each outcome. Our findings suggest that HTN-AI does not only identify patients at risk of hypertension, but also of downstream sequelae of hypertension.
To understand whether the HTN-AI score provides additional information about cardiovascular risk in patients with known hypertension, we examined CVD incidence stratified by both HTN-AI risk and baseline hypertension status. HTN-AI risk stratified cumulative incidence of mortality among both patients with and without a baseline diagnosis of hypertension (Fig. 3a and Supplementary Table 6). Among 30,771 patients with known hypertension, 10,627 had high predicted HTN-AI risk and 20,144 had low predicted HTN-AI risk. Over a median follow up time of 5.7 years, there were 3480/10,627 (32.7%) deaths in the high HTN-AI risk group and 2622/20,144 (13.0%) deaths in the low HTN-AI risk group (estimated 10-year cumulative incidence 39.9% [38.7–41.1] for high HTN-AI risk vs 13.8% [13.3–14.4] for low HTN-AI risk). The pattern was similar among patients without baseline hypertension and for outcomes other than mortality (Supplementary Table 6).
The figure depicts cumulative incidence of all-cause mortality stratified by a HTN-AI risk and baseline hypertension status, b HTN-AI risk and age, and c quintile of HTN-AI risk. a To determine the additional clinical utility of HTN-AI over baseline hypertension status, we assessed cumulative incidence of cardiovascular outcomes stratified by both HTN-AI risk and baseline hypertension status. High HTN-AI risk was associated with increased risk of adverse outcomes regardless of baseline hypertension status, suggesting that HTN-AI detects electrocardiographic features of hypertension that are relevant to downstream risk of adverse sequelae of hypertension. b We also assessed cumulative incidence of cardiovascular outcomes stratified by age, demonstrating that the HTN-AI score stratified risk of adverse outcomes independent of age. c We stratified cumulative incidence of cardiovascular outcomes by quintile of HTN-AI score. The HTN-AI score represents a gradient of cardiovascular risk that is most concentrated in the top quintile, suggesting that the HTN-AI score may be especially useful to identify patients at the highest risk of downstream cardiovascular disease.
We also examined cumulative incidence of CVD stratified by HTN-AI risk and age, finding that HTN-AI risk stratified cumulative incidence of all outcomes within age strata (Fig. 3b and Supplementary Table 7). We additionally stratified cumulative incidence by the quintile of HTN-AI score, finding that events were concentrated among patients in the top quintile with a gradient of risk at lower quintiles (Fig. 3c, and Supplementary Table 8).
In regression models accounting for competing risk of death, the HTN-AI score was significantly associated with mortality (HR per SD: 1.47 [1.36–1.60], p < 0.001), HF (2.26 [1.90–2.69], p < 0.001), MI (1.87 [1.69–2.07], p < 0.001), stroke (1.30 [1.18–1.44], p < 0.001), and aortic dissection or rupture (1.69 [1.22–2.35], p < 0.001), after adjustment for age, sex, baseline hypertension status, baseline hypertension medication use, diabetes, hyperlipidemia, body mass index, and current smoking status. Figure 4 compares adjusted HRs per SD of HTN-AI score, SBP, and pulse pressure, demonstrating that effect sizes were higher for HTN-AI than for SBP or pulse pressure across all outcomes.
To compare the associations of blood pressure measurements and the HTN-AI score with incidence cardiovascular outcomes, we performed cause-specific hazard regression of five cardiovascular outcomes (mortality, heart failure, myocardial infarction, ischemic stroke, and aortic dissection or rupture) against each of HTN-AI, baseline systolic blood pressure, and baseline pulse pressure, with adjustment for age, sex, baseline hypertension status, baseline hypertension medication use, diabetes, hyperlipidemia, body mass index, and current smoking status. Pulse pressure was included in place of diastolic blood pressure due to the non-linear relationship between diastolic blood pressure and age. The figure depicts a forest plot of the results, with each hazard ratio representing the adjusted subdistribution hazard ratio per standard deviation of the covariate. Error bars represent 95% confidence intervals. Across all outcomes, the effect size per standard deviation of the HTN-AI score is greater than the corresponding effect size per standard deviation of each blood pressure metric. Our results suggest that the HTN-AI score captures more information about risk of hypertension-associated cardiovascular disease than does office blood pressure measurement. BP: blood pressure. HR: hazard ratio. SD: standard deviation.
Finally, we assessed discriminative performance of the HTN-AI score as a digital biomarker for incident CVD using Harrell’s C-statistic (Supplementary Table 9). The HTN-AI score significantly discriminated incidence of all CVD outcomes (C-statistic range 0.704–0.804). Discrimination for the HTN-AI score was higher than the discrimination of age for HF (HTN-AI score C-statistic 0.804 [0.796–0.813] vs age C-statistic 0.725 [0.713–0.736]) and MI (HTN-AI score C-statistic 0.733 [0.725–0.742] vs age C-statistic 0.665 [0.656–0.675]). We also compared discrimination of the HTN-AI score to discrimination of established clinical risk scores for HF and atherosclerotic CVD. HTN-AI discrimination of HF was comparable to the Pooled Cohort Equation to Prevent Heart Failure (PCP-HF) score16 (HTN-AI score C-statistic 0.804 [0.796–0.813] vs PCP-HF C-statistic 0.806 [0.773–0.839]). HTN-AI also demonstrated superior discrimination compared the Pooled Cohort Equation (PCE)17 for MI (HTN-AI score C-statistic 0.733 [0.725–0.742] vs PCE C-statistic 0.681 [0.670–0.693]) and stroke (HTN-AI score C-statistic 0.704 [0.693–0.714] vs PCE C-statistic 0.683 [0.669–0.698]).
Model interpretability
To understand how HTN-AI is influenced by standard ECG measurements, we examined the proportion of variability in the HTN-AI score explained by ECG measurements among the 56,760 patients in the BWH external validation sample using linear regression. The examined ECG measurements included the heart rate; PR, QRS, and QT intervals; P-wave, R-wave, and T-wave axes; S-wave amplitudes in leads V1 and V3; and R-wave amplitudes in aVL, V5, and V6. The greatest variability in HTN-AI score was explained by the R wave amplitude in aVL (R2 = 0.175), R-wave axis (R2 = 0.163), and QRS interval (R2 = 0.077). The adjusted R2 for a multivariable model including all ECG measurements was 0.358. Intuitively, longer PR, QRS and QT intervals, higher aVL R-wave amplitude, and more leftward R-wave axis were associated with higher HTN-AI scores (Supplementary Table 10). However, we also noted that lower V1 S amplitude, V5 R amplitude, and V6 R amplitude were associated with higher HTN-AI scores (Supplementary Table 10), suggesting that HTN-AI predictions do not correlate linearly with conventional voltage criteria for left ventricular hypertrophy.
We also sought to explain HTN-AI predictions by generating saliency maps and median ECG waveforms for 1024 randomly selected patients from the top and bottom deciles of HTN-AI score in the BWH sample (Supplementary Fig. 4,5) to visualize which areas of the ECG contribute to HTN-AI predictions. Saliency maps highlight regions of the ECG that are influential on model outputs, and qualitatively demonstrated that the most salient regions of the ECG vary from lead to lead, though generally areas of the ECG that do not correspond to atrial or ventricular depolarization or repolarization (i.e. the T–P segment) do not contribute significantly to model predictions. Qualitative comparison of median waveforms of high and low risk patients demonstrates that higher voltages, wider QRS complexes, and wider and flatter P- and T-waves are associated with higher HTN-AI scores.
Finally, we also assessed whether associations between the HTN-AI score and CVD are sensitive to presence of other ECG abnormalities by examining the associations between HTN-AI score and incident CVD in the subset of patients with ECGs interpreted by a cardiologist as normal. Among patients in the BWH sample with normal ECGs, the association with the HTN-AI score remained significant for all outcomes except aortic dissection or rupture (p = 0.300; p < 0.05 for other outcomes; Supplementary Fig. 6), suggesting that HTN-AI retains prognostic value even when there are no overt ECG abnormalities.
Discussion
Here we present HTN-AI, a deep learning-based digital biomarker that was trained to identify hypertension exclusively from signals contained within the 12-lead ECG waveform. HTN-AI was trained using over 750,000 ECGs from over 100,000 patients, and externally validated in a sample of over 56,000 patients from a second hospital. In this work we demonstrate that HTN-AI accurately identifies prevalent hypertension, stratifies risk of short-term incidence of hypertension diagnosis, and is associated with elevated BP as measured by gold-standard 24-hour ambulatory BP monitoring. We further demonstrate that HTN-AI stratifies risk of hypertension-associated CVD regardless of baseline hypertension status or presence of overt ECG abnormalities, suggesting that it may be useful as a digital biomarker of hypertension-associated cardiovascular risk.
Deep learning applied to 12-lead ECGs has previously been used to classify prevalent18 and incident19 atrial fibrillation, left ventricular systolic dysfunction20, valvular disease21, and mortality22, among other CVD states, demonstrating the feasibility of disease prediction from 12-lead ECG signals. While small non-clinical studies23,24 have used deep learning to classify prevalent hypertension from ECGs, our work does so at scale using clinical data and demonstrates the associations of model-predicted probability of hypertension with incident CVD.
Office BP is often used to diagnose and manage hypertension but does not always reflect ambulatory BP, leading to potential underdiagnosis of ambulatory hypertension1,10. In contrast, HTN-AI detects electrocardiographic features of hypertension which result from the cumulative effect of high ambulatory BP on the myocardium, which is well-known to cause changes in cardiac structure and conduction. HTN-AI may therefore be useful to screen for ambulatory hypertension even when office BP is normal by identifying patients with subclinical ECG features of hypertension which could prompt confirmatory testing with ambulatory BP monitoring.
As an example, HTN-AI could be deployed to opportunistically screen for masked hypertension in the primary care setting using the 12-lead ECG. Patients without hypertension but with high ECG-based HTN-AI risk could be screened with 24-hour ABPM. Assuming a 10% prevalence of masked hypertension as previously estimated1, HTN-AI would have an estimated positive predictive value of approximately 0.3 based on our validation sensitivity and specificity, suggesting that for every 3-4 high HTN-AI risk patients screened with ABPM 1 would be diagnosed with hypertension. However, further work is needed to validate HTN-AI specifically for detection of masked hypertension.
Though HTN-AI was only trained to identify hypertension from the ECG it also stratifies risk of hypertension-related CVD and does so regardless of baseline hypertension status. Our findings suggest that HTN-AI can serve as a digital biomarker for hypertension-associated cardiovascular risk by detecting electrocardiographic features of hypertension that are also associated with incident CVD. In fact, we observed greater normalized effect sizes for the HTN-AI score than for baseline BP across all CVD outcomes, suggesting that HTN-AI is a better marker of cardiovascular risk than convenience office BP measurements. Additionally, for HF and MI the HTN-AI score achieved comparable or superior discrimination to established clinical risk scores without requiring any prior knowledge about the patient aside from a 12-lead ECG. Notably, it remains unknown whether the HTN-AI score represents accumulated non-modifiable cardiovascular damage or modifiable risk that can be reversed with treatment.
Deep learning methods are well-suited to learning from high-dimensional inputs such as the ECG. We demonstrate that despite significant correlations between the HTN-AI score and standard ECG measurements, when combined these measurements still only explain a fraction of the variability captured by our model, which is able to learn highly non-linear interactions between various regions of the ECG waveform. However, the strong performance afforded by the non-linearity of deep learning methods comes at the cost of limited interpretability. To mitigate this limitation, we provide saliency maps and median waveforms that suggest HTN-AI focuses on ECG regions that are known to exhibit hypertension-related changes. Furthermore, the strong associations between HTN-AI predictions and CVD suggest that the model detects biologically relevant signal in ECG waveforms that correlate with the physiological effects of hypertension.
Our study should be interpreted in the context of its limitations. Given the size of our sample which exceeded 150,000 patients, hypertension status for model training was by necessity determined using office BPs and diagnosis codes rather than gold-standard ambulatory BP monitoring, creating potential for misclassification. However, we minimized misclassification by employing study cohorts that were purpose-built to limit missingness and ascertainment bias, and our outcomes cohort was previously validated with respect to established cardiovascular risk models including BP measurements25. Furthermore, the scale of our training sample allows the model to identify electrocardiographic patterns associated with hypertension aggregated across 750,000 training ECGs, despite the noise introduced by potential misclassification of individual patients. We also compare the HTN-AI score to gold-standard 24-hour ambulatory BP monitoring, showing that high HTN-AI risk is associated with higher mean 24-hour systolic BP and greater odds of abnormal 24-hour ambulatory BP monitoring results. Finally, HTN-AI predictions are strongly associated with incident CVD, indicating that the model identifies a physiological ECG signature of hypertension despite learning from training labels derived from electronic health records. Nevertheless, prior to implementation in clinical practice it will be necessary to prospectively validate the HTN-AI score against gold-standard measures of hypertension such as 24-hour ABPM. Additionally, though we trained and validated our model in distinct populations from 2 hospitals, both are large academic centers within one hospital system representing a single geographic region of the United States.
HTN-AI is a deep learning model that identifies hypertension from 12-lead ECG signals and stratifies risk of incident mortality, HF, MI, stroke, and aortic dissection or rupture. Our work demonstrates the potential of a deep learning model to facilitate the diagnosis of hypertension and serve as a novel digital biomarker for the risk of hypertension-associated cardiovascular disease.
Methods
Study population
The study protocol was approved by the MGH Institutional Review Board (Protocol #2017P001650) and complied with the Declaration of Helsinki. The requirement for informed consent was waived by the institutional review board given the low-risk retrospective nature of the study and privacy-protecting measures implemented by the investigators. The study samples were derived from 2 longitudinal electronic health record (EHR)-based cohorts of ambulatory patients within the Mass General Brigham (MGB) healthcare system (Fig. 1). The first cohort was the Community Care Cohort Project, a cohort of 520,868 patients aged 18 to 89 years with multiple visits to MGB primary care clinics between 2001 and 2018. The cohort was designed to ensure accurate ascertainment of baseline comorbidities by capturing only patients receiving longitudinal primary care within the MGB system, and to minimize missingness by using natural language processing to recover missing data from clinical notes25. Comparison of the Community Care Cohort Project to a convenience EHR sample demonstrated better calibration for established risk models including the Pooled Cohort Equation and Cohorts for Heart and Aging Research in Genomic Epidemiology Atrial Fibrillation (CHARGE-AF) risk score25. The second cohort was the Enterprise Warehouse of Cardiology, an analogously designed cohort of 99,252 longitudinal cardiology patients between the ages of 18 and 89 years with multiple visits to MGB cardiology clinics between 2000 and 201926. Start of follow up in each cohort was the date of the second clinic visit that determined cohort inclusion.
The HTN-AI model was trained in patients in either cohort with 1 or more 12-lead ECG performed at MGH within 3 years of the start of follow up. Internal validity was assessed in a hold-out MGH validation sample using the most recent ECG performed within the 3 years before the start of follow up. Patients missing data for age or sex were excluded. External validation and time-to-event analyzes were performed in patients in the primary care cohort with 12-lead ECGs done at BWH within the 3 years before the start of follow up (BWH external validation sample). Patients with both MGH and BWH ECGs were included only in the training sample, and patients missing data for age, date of last follow up, or sex were excluded.
Patient characteristics and outcomes
Age, sex, race, ethnicity, comorbidities, and outcomes were determined using EHR data. Race and ethnicity were extracted from a combined EHR field. Baseline comorbidities, including atrial fibrillation, chronic kidney disease, coronary artery disease, diabetes mellitus, HF, and hypertension, were determined by presence of a single International Classification of Diseases (ICD)-9 or ICD-10 code corresponding to the comorbidity (see Supplementary Table 1) by the start of cohort follow up, except for atrial fibrillation and HF. Atrial fibrillation was defined using a previously published algorithm that uses diagnostic and procedural codes and ECG reports27. HF was defined using diagnosis codes for a primary diagnosis of HF for inpatient encounters. Hypertension medication use was determined from EHR medication records. Baseline BP, body mass index, and smoking status was defined using the most recent values within the 3 years prior to start follow up and was obtained using structured EHR data and natural language processing of clinical notes25. Body mass index was filtered to a range of 5 kg/m2 to 150 kg/m2. If body mass index data was unavailable, it was calculated from height and weight data, if available. Height was filtered to a range of 121.92 cm (4 feet) to 304.8 cm (10 feet). Weight was filtered to a range of 13.6 kg (30 lbs) to 454.5 kg (1000 lbs). Within the BWH validation sample, we identified patients who underwent 24-hour ambulatory BP monitoring using Current Procedural Terminology codes (93784, 93786, 93788, 93790, and A4670) and manually reviewed ambulatory BP monitoring results to obtain 24-hour average systolic and diastolic BP.
CVD outcomes included all-cause mortality, HF, MI, stroke, and aortic dissection or rupture. Dates of death were determined using EHR records. Dates of incident HF were defined as the date of the first inpatient diagnosis code for HF in the primary position. Dates of incidence of other outcomes were the date of the first corresponding diagnosis code after the start of follow up.
Electrocardiogram acquisition and preprocessing
ECGs and metadata were obtained from the MUSE Cardiology Information System (GE Healthcare). Prior to presentation to the model, voltages for each ECG are normalized to have mean zero and standard deviation of 1. ECGs recorded at 250 Hz are bilinearly upsampled to 500 Hz and leads with missing data are zero-padded such that all 12 leads contain 5000 voltages. ECGs without abnormalities were identified via text matching using physician interpretations.
Model development and training
Patients were considered to have prevalent hypertension if they had (1) a baseline diagnosis of hypertension by diagnosis code, (2) baseline SBP ≥ 140 mmHg, if available, or (3) baseline DBP ≥ 90 mmHg, if available. Multiple ECGs were used per patient during training including ECGs before and after the start of follow up. During validation and time-to-event analyzes, only the most recent ECG before the start of follow up was used. Training, development, and internal validation samples were generated using a 70/15/15 percent split at the patient level.
The HTN-AI model takes as input a 12-lead ECG waveform and provides 4 outputs: probability of hypertension, probability of antihypertensive medication use, probability of male sex, and estimated age. No clinical risk factors or non-ECG data is provided to the model. The model architecture consists of a series of 1-dimensional convolutions followed by nonlinear activation functions and downsampling operations which transform the input from a 5000×12 tensor (i.e. a 10 second ECG sampled at 500 Hz with 12 leads) to a 256-dimension embedding, which is used by output layers to generate predictions. The cross entropy loss for the categorical tasks and the logarithmic hyperbolic cosine loss for age regression were summed and minimized using the Adam stochastic gradient descent algorithm28.
Model hyper-parameters including depth, width, convolutional kernel size, and activation function were selected through Bayesian hyperparameter optimization. The resulting architecture contains 15,072,839 parameters organized into 3 blocks of densely-connected parallel convolutions with kernel size of 71, which pass through the swish activation function and are merged together and down-sampled by max-pooling, as in the Densenet architecture29,30. A dropout rate of 0.5 on dense layers and spatial dropout rate of 0.2 on convolutional layers are used during training to stochastically sample from the space of architectures and mitigate overfitting30,31. Weights are updated using the Adam optimizer with batch size of 24, initial learning rate of 5e-4. The learning rate decays by a factor of 0.5 after 32 epochs without an improvement in validation loss32. Early stopping is used to checkpoint the model parameters with minimum validation loss during training, and it is those model parameters which are used for all subsequent evaluation. Training takes approximately 5 hours until convergence on a NVidia V-100 GPU.
Deep learning model sensitivity analysis
To better understand what drives the performance of HTN-AI we conducted ablation studies using different definitions of hypertension and different sets of auxiliary tasks in model training. The primary model defined prevalent hypertension as any combination of (1) a baseline diagnosis of hypertension by ICD-9 or ICD-10 codes, (2) baseline SBP ≥140 mmHg, or (3) baseline DBP ≥ 90 mmHg. The primary model was trained using this hypertension label with auxiliary tasks of age regression, sex classification, and classification of baseline antihypertensive medication use. We tested three other combinations of hypertension definitions and tasks:
-
A single-task model using the same definition of prevalent hypertension without simultaneous age regression, sex classification, or classification of baseline antihypertensive medication use.
-
A single-task model that also incorporated use of anti-hypertensive medication into the definition of prevalent hypertension. For this model anti-hypertensive medication use was not used as an auxiliary training task.
-
A single-task model that included only ICD-9 or ICD-10 code diagnosis of hypertension without consideration of baseline BP.
For models that incorporated baseline BP measurements, if baseline BP measurements were not available they were not used in determining the training label. Supplementary Table 2 details performance of the primary model and sensitivity analysis models in both the MGH and BWH test samples.
Statistical analyses
We report normally distributed data as means (standard deviation [SD]), non-normally distributed data as median (interquartile range) and categorical data as counts (%). High and low HTN-AI risk groups were determined using a HTN-AI score cutoff of 0.85, defined to achieve 90% specificity in the model development sample. Model performance for hypertension classification is reported as area under the receiver operating characteristicor curve (AUROC), sensitivity/recall, specificity, precision, average precision, and F1 score.
We tested the association between HTN-AI risk group and elevated 24-hour ambulatory BP using logistic regression with adjustment for age, sex, and use of an antihypertensive medication prior to ambulatory BP monitoring and report 2-sided Wald p-values. For incident disease analyzes, we show cause-specific cumulative incidence curves for each outcome with 2-sided p-values for equivalence between groups calculated by Gray’s test, accounting for competing risk of death for outcomes other than mortality. Cause-specific Fine-Gray regression was performed accounting for competing risk of death for outcomes other than mortality, with adjustment for age, sex, body mass index, and baseline hypertension, diabetes mellitus, hyperlipidemia, smoking status, and antihypertensive medication use. Subdistribution hazard ratios (HRs) and 95% confidence intervals with 2-sided p-values are reported. Patients were right-censored at the last EHR encounter or on 8/31/19. We included only cases with complete covariate data and with available BP data (n = 23,705 [41.8%]) to facilitate comparison between models using HTN-AI score, SBP and pulse pressure. To assess discrimination of the HTN-AI score for incident CVD we calculated Harrell’s C-statistic. We assessed associations between the HTN-AI score and standard ECG measurements using linear regression and report beta statistics, 95% confidence intervals, and univariable and adjusted multivariable R2.
We also performed a sensitivity analysis examining the association of HTN-AI with incident cardiovascular disease in patients with normal ECGs, to assess whether the association is sensitive to the presence of abnormal ECG findings. We obtained the text of cardiologist ECG reads from the MUSE ECG database. The text of cardiologist ECG reads was searched for the phrase “Normal Sinus Rhythm Normal ECG” which is a common phrase, entered using a text macro, used by cardiologists in our system for ECGs that contain no abnormalities. This was intended to be a specific rather than sensitive search for ECGs without clinically significant abnormal features. ECGs in the BWH test sample that contained the phrase were considered normal, and this subset of ECGs was used in an identical cause-specific cumulative incidence regression as described above. The results are depicted in Supplementary Fig. 6.
Data availability
MGB patient data is protected health information and is unable to be shared.
Code availability
The model was implemented with the ML4H Python library33 (version 0.0.4) using the Tensorflow34 (version 2.9.1) framework. The ML4H Python library is available at https://github.com/broadinstitute/ml4h. Statistical analyzes were done using R (version 4.2.1) with packages cmprsk (version 2.2-11), survival (version 3.4-0), survminer (version 0.4.9), and yardstick (version 1.1.0).
References
Unger, T. et al. 2020 International Society of Hypertension Global Hypertension Practice Guidelines. Hypertension 75, 1334–1357 (2020).
Bundy, J. D. et al. Systolic blood pressure reduction and risk of cardiovascular disease and mortality: a systematic review and network meta-analysis. JAMA Cardiol 2, 775–781 (2017).
Rapsomaniki, E. et al. Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1·25 million people. Lancet 383, 1899–1911 (2014).
Pencina, M. J. et al. Quantifying importance of major risk factors for coronary heart disease. Circulation 139, 1603–1611 (2019).
Sinha, A. et al. Race- and sex-specific population attributable fractions of incident heart failure. Circ. Heart Fail. 14, e008113 (2021).
Patel, S. A., Winkel, M., Ali, M. K., Narayan, K. M. V. & Mehta, N. K. Cardiovascular mortality associated with 5 leading risk factors: national and state preventable fractions estimated from survey data. Ann. Intern. Med. 163, 245–253 (2015).
Tsao, C. W. et al. Heart Disease and Stroke Statistics-2023 Update: A Report From the American Heart Association. Circulation 147, e93–e621 (2023).
Whelton, P. K. et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 71, e13–e115 (2018).
Stergiou, G. S. et al. European Society of Hypertension practice guidelines for office and out-of-office blood pressure measurement. J. Hypertens. 39, 1293–1302 (2021).
Lu, Y. et al. Quantifying blood pressure visit-to-visit variability in the real-world setting: a retrospective cohort study. Circ. Cardiovasc. Qual. Outcomes 16, e009258 (2023).
Hwang, K. O., Aigbe, A., Ju, H.-H., Jackson, V. C. & Sedlock, E. W. Barriers to Accurate Blood Pressure Measurement in the Medical Office. J. Prim. Care Community Health 9, 2150132718816929 (2018).
Booth, J. N. 3rd et al. Masked hypertension and cardiovascular disease events in a prospective cohort of blacks: the Jackson heart study. Hypertension 68, 501–510 (2016).
Shimbo, D. et al. The use of ambulatory blood pressure monitoring among Medicare beneficiaries in 2007-2010. J. Am. Soc. Hypertens. 8, 891–897 (2014).
Lehtonen, A. O. et al. Prevalence and prognosis of ECG abnormalities in normotensive and hypertensive individuals. J. Hypertens. 34, 959–966 (2016).
Coravos, A., Khozin, S. & Mandl, K. D. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. NPJ Digit Med. 2, 14 (2019).
Khan, S. S. et al. 10-year risk equations for incident heart failure in the general population. J. Am. Coll. Cardiol. 73, 2388–2397 (2019).
Goff, D. C. Jr et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 129, S49–S73 (2014).
Attia, Z. I. et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 394, 861–867 (09/2019).
Khurshid, S. et al. ECG-based deep learning and clinical risk factors to predict atrial fibrillation. Circulation 145, 122–133 (2022).
Attia, Z. I. et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 25, 70–74 (2019).
Elias, P. et al. Deep Learning Electrocardiographic Analysis for Detection of Left-Sided Valvular Heart Disease. J. Am. Coll. Cardiol. 80, 613–626 (2022).
Raghunath, S. et al. Deep neural networks can predict new-onset atrial fibrillation from the 12-lead ecg and help identify those at risk of atrial fibrillation-related stroke. Circulation 143, 1287–1298 (2021).
Jain, P., Gajbhiye, P., Tripathy, R. K. & Acharya, U. R. A two-stage deep CNN architecture for the classification of low-risk and high-risk hypertension classes using multi-lead ECG signals. Informatics Medicine Unlocked 21, 100479 (2020).
Soh, D. C. K. et al. Automated diagnostic tool for hypertension using convolutional neural network. Comput. Biol. Med. 126, 103999 (2020).
Khurshid, S. et al. Cohort design and natural language processing to reduce bias in electronic health records research. NPJ Digit. Med. 5, 47 (2022).
Singh, P. et al. One clinician is all you need-cardiac magnetic resonance imaging measurement extraction: deep learning algorithm development. JMIR Med. Inform. 10, e38178 (2022).
Khurshid, S., Keaney, J., Ellinor, P. T. & Lubitz, S. A. A simple and portable algorithm for identifying atrial fibrillation in the electronic medical record. Am. J. Cardiol. 117, 221–225 (2016).
Kingma et al. A method for stochastic optimization. https://doi.org/10.48550/arXiv.1412.6980 (2014)
Iandola, F. et al. DenseNet: Implementing efficient ConvNet descriptor pyramids. arXiv [cs.CV] (2014).
Ramachandran, P., Zoph, B. & Le, Q. V. Searching for Activation Functions. arXiv [cs.NE] (2017).
Park, S. & Kwak, N. Analysis on the dropout effect in convolutional neural networks. In Computer Vision – ACCV 2016 189–204 (Springer International Publishing, Cham, https://doi.org/10.1007/978-3-319-54184-6_12. 2017).
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization. arXiv [cs.LG] (2014).
Ml4h. (Github).
Abadi, M. et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv [cs.DC] (2016).
Acknowledgements
MAA is supported by a grant from the National Institutes of Health (NIH; T32-HL007208). SKa is supported by the Walter Benjamin Fellowship from the Deutsche Forschungsgemeinschaft (521832260). JTR is supported by a Fellowship grant from the Sigrid Jusélius Foundation. SKh is supported by a grant from the NIH (K23HL169839-01) and American Heart Association (AHA; 2023CDA1050571). JPP is supported by a grant from the NIH (K08HL159346). JEH is supported by grants from the NIH (R01HL134893, R01HL140224, R01HL160003, K24HL153669). PTE is supported by grants from the NIH (1R01HL092577, 1R01HL157635), by a grant from the AHA (18SFRN34230127, 961045), and from the European Union (MAESTRIA 965286).
Author information
Authors and Affiliations
Contributions
M.A.A. and S.F.F. contributed equally to this manuscript. M.A.A., and S.F.F. designed the study. M.A.A. additionally performed statistical analyzes, interpreted and visualized results, and wrote the original draft of the manuscript. S.F.F. additionally developed the deep learning model and the underlying software used to create it, and contributed to writing the original manuscript draft. S.Ka., J.T.R., D.P., S.Kh., and J.P.P. contributed to study design and interpretation of data, and reviewed and edited the manuscript for critical intellectual content. P.S. and C.R. developed the data processing pipeline and performed data curation and validation. M.M., J.E.H., and P.T.E. supervised the study and reviewed and edited the manuscript for critical intellectual content. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
MAA, SFF, and PTE are coinventors on US Provisional Patent Application No. 63/716,507 filed by applicants The Broad Institute, Inc. and The General Hospital Corporation, covering all aspects of the work described in this manuscript. SKh receives sponsored research support from Bayer AG. JEH has previously received sponsored research support from Bayer AG. PTE receives sponsored research support from Bayer AG, Bristol Myers Squibb, Pfizer, and Novo Nordisk; he has also served on advisory boards or consulted for Bayer AG. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Al-Alusi, M.A., Friedman, S.F., Kany, S. et al. A deep learning digital biomarker to detect hypertension and stratify cardiovascular risk from the electrocardiogram. npj Digit. Med. 8, 120 (2025). https://doi.org/10.1038/s41746-025-01491-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-025-01491-8