Abstract
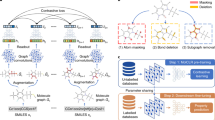
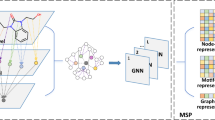
Successful deep learning critically depends on the representation of the learned objects. Recent state-of-the-art pharmaceutical deep learning models successfully exploit graph-based de novo learning of molecular representations. Nonetheless, the combined potential of human expert knowledge of molecular representations and convolution neural networks has not been adequately explored for enhanced learning of pharmaceutical properties. Here we show that broader exploration of human-knowledge-based molecular representations enables more enhanced deep learning of pharmaceutical properties. By broad learning of 1,456 molecular descriptors and 16,204 fingerprint features of 8,506,205 molecules, a new feature-generation method MolMap was developed for mapping these molecular descriptors and fingerprint features into robust two-dimensional feature maps. Convolution-neural-network-based MolMapNet models were constructed for out-of-the-box deep learning of pharmaceutical properties, which outperformed the graph-based and other established models on most of the 26 pharmaceutically relevant benchmark datasets and a novel dataset. The MolMapNet learned important features that are consistent with the literature-reported molecular features.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The full datasets and corresponding annotations are available on GitHub at https://github.com/shenwanxiang/ChemBench/tree/v0 and on Zenodo at https://doi.org/10.5281/zenodo.405486671. Source data are provided with this paper.
Code availability
Codes for the MolMap and MolMapNet package and the parameters are available on GitHub and CodeOcean, together with the data used for testing the package, at https://github.com/shenwanxiang/bidd-molmap and https://codeocean.com/capsule/2307823/tree72.
References
Paolini, G. V., Shapland, R. H. B., van Hoorn, W. P., Mason, J. S. & Hopkins, A. L. Global mapping of pharmacological space. Nat. Biotechnol. 24, 805–815 (2006).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Xiong, Z. et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J. Med. Chem. 63, 8749–8760 (2019).
Yang, K. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019).
Duvenaud, D. K. et al. Convolutional networks on graphs for learning molecular fingerprints. Adv. Neural Inf. Process. Syst. 28, 2224–2232 (2015).
Moriwaki, H., Tian, Y. S., Kawashita, N. & Takagi, T. Mordred: a molecular descriptor calculator. J. Cheminform. 10, 4 (2018).
Landrum, G. RDKit Documentation Release 2019.09.1, 1-151 http://www.rdkit.org (2019).
Bengio, Y., Courville, A. & Vincent, P. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828 (2013).
Chen, Y., Bai, Y., Zhang, W. & Mei, T. Destruction and construction learning for fine-grained image recognition. In IEEE Conference on Computer Vision and Pattern Recognition 5157–5166 (CVPR, 2019).
Johnson, R. & Zhang, T. Effective use of word order for text categorization with convolutional neural networks. In Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 103–112 (ACL, 2014).
Han, S., Pool, J., Tran, J. & Dally, W. Learning both weights and connections for efficient neural network. In Proceedings of Advances in Neural Information Processing Systems 1135–1143 (NIPS, 2015).
Dong, J. et al. PyBioMed: a Python library for various molecular representations of chemicals, proteins and DNAs and their interactions. J. Cheminform. 10, 16 (2018).
Winter, R., Montanari, F., Noé, F. & Clevert, D.-A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 10, 1692–1701 (2019).
Probst, D. & Reymond, J.-L. A probabilistic molecular fingerprint for big data settings. J. Cheminform. 10, 66 (2018).
Allen, C. H. G., Mervin, L. H., Mahmoud, S. Y. & Bender, A. Leveraging heterogeneous data from GHS toxicity annotations, molecular and protein target descriptors and Tox21 assay readouts to predict and rationalise acute toxicity. J. Cheminform. 11, 36 (2019).
Clemons, P. A. et al. Quantifying structure and performance diversity for sets of small molecules comprising small-molecule screening collections. Proc. Natl Acad. Sci. USA 108, 6817–6822 (2011).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Yao, Q. et al. Taking human out of learning applications: a survey on automated machine learning. Preprint at https://arxiv.org/abs/1810.13306 (2018).
Sun, M. et al. Graph convolutional networks for computational drug development and discovery. Brief. Bioinform. 21, 919–935 (2019).
Popova, M., Isayev, O. & Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 4, eaap7885 (2018).
Goh, G. B., Hodas, N. O., Siegel, C. & Vishnu, A. Smiles2vec: an interpretable general-purpose deep neural network for predicting chemical properties. Preprint at https://arxiv.org/abs/1712.02034 (2017).
Karpov, P., Godin, G. & Tetko, I. V. Transformer-CNN: Swiss knife for QSAR modeling and interpretation. J. Cheminform. 12, 17 (2020).
Goh, G. B., Siegel, C., Vishnu, A. & Hodas, N. O. Chemnet: a transferable and generalizable deep neural network for small-molecule property prediction. Preprint at https://arxiv.org/abs/1712.02734 (2017).
Cortés-Ciriano, I. & Bender, A. KekuleScope: prediction of cancer cell line sensitivity and compound potency using convolutional neural networks trained on compound images. J. Cheminform. 11, 41 (2019).
Wenzel, J., Matter, H. & Schmidt, F. Predictive multitask deep neural network models for ADME-Tox properties: learning from large data sets. J. Chem. Inf. Model. 59, 1253–1268 (2019).
Ivan, C. Convolutional neural networks on randomized data. In IEEE Conference on Computer Vision and Pattern Recognition Workshops 1–8 (CVPR, 2019).
Bajusz, D., Rácz, A. & Héberger, K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 7, 20 (2015).
Pyka, A., Babuska, M. & Zachariasz, M. A comparison of theoretical methods of calculation of partition coefficients for selected drugs. Acta Pol. Pharm. 63, 159–167 (2006).
Li, X., Xu, Y., Lai, L. & Pei, J. Prediction of human cytochrome P450 inhibition using a multitask deep autoencoder neural network. Mol. Pharm. 15, 4336–4345 (2018).
Ramsundar, B. et al. Massively multitask networks for drug discovery. Preprint at https://arxiv.org/abs/1502.02072 (2015).
Peng, C., Zhang, X., Yu, G., Luo, G. & Sun, J. Large Kernel matters—improve semantic segmentation by global convolutional network. In IEEE Conference on Computer Vision and Pattern Recognition 4353–4361 (CVPR, 2017).
Bento, A. P. et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 42, D1083–D1090 (2014).
Probst, D. & Reymond, J.-L. Visualization of very large high-dimensional data sets as minimum spanning trees. J. Cheminform. 12, 12 (2020).
Godden, J. W., Stahura, F. L. & Bajorath, J. Anatomy of fingerprint search calculations on structurally diverse sets of active compounds. J. Chem. Inf. Model. 45, 1812–1819 (2005).
Fisher, A., Rudin, C. & Dominici, F. All models are wrong, but many are useful: learning a variable’s importance by studying an entire class of prediction models simultaneously. J. Mach. Learn. Res. 20, 1–81 (2019).
Huuskonen, J. Estimation of aqueous solubility for a diverse set of organic compounds based on molecular topology. J. Chem. Inf. Comput. Sci. 40, 773–777 (2000).
Tetko, I. V., Tanchuk, V. Y., Kasheva, T. N. & Villa, A. E. P. Estimation of aqueous solubility of chemical compounds using E-state indices. J. Chem. Inf. Comput. Sci. 41, 1488–1493 (2001).
Huuskonen, J., Rantanen, J. & Livingstone, D. Prediction of aqueous solubility for a diverse set of organic compounds based on atom-type electrotopological state indices. Eur. J. Med. Chem. 35, 1081–1088 (2000).
Huuskonen, J. Estimation of water solubility from atom‐type electrotopological state indices. Environ. Toxicol. Chem. 20, 491–497 (2001).
Ensing, B. et al. On the origin of the extremely different solubilities of polyethers in water. Nat. Commun. 10, 2893 (2019).
Cheng, Y. et al. From fragment screening to in vivo efficacy: optimization of a series of 2-aminoquinolines as potent inhibitors of beta-site amyloid precursor protein cleaving enzyme 1 (BACE1). J. Med. Chem. 54, 5836–5857 (2011).
Madden, J. et al. Fragment-based discovery and optimization of BACE1 inhibitors. Bioorg. Med. Chem. Lett. 20, 5329–5333 (2010).
Van De Waterbeemd, H. & Gifford, E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2, 192–204 (2003).
Altae-Tran, H., Ramsundar, B., Pappu, A. S. & Pande, V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017).
Glavatskikh, M., Leguy, J., Hunault, G., Cauchy, T. & Da Mota, B. Dataset’s chemical diversity limits the generalizability of machine learning predictions. J. Cheminform. 11, 69 (2019).
Townsend, J., Micucci, C. P., Hymel, J. H., Maroulas, V. & Vogiatzis, K. D. Representation of molecular structures with persistent homology for machine learning applications in chemistry. Nat. Commun. 11, 3230 (2020).
Kim, S. et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Gedeck, P., Rohde, B. & Bartels, C. QSAR—How good is it in practice? Comparison of descriptor sets on an unbiased cross section of corporate data sets. J. Chem. Inf. Model. 46, 1924–1936 (2006).
Capecchi, A., Probst, D. & Reymond, J.-L. One molecular fingerprint to rule them all: drugs biomolecules, and the metabolome. J. Cheminform. 12, 43 (2020).
Stiefl, N., Watson, I. A., Baumann, K. & Zaliani, A. ErG: 2D pharmacophore descriptions for scaffold hopping. J. Chem. Inf. Model. 46, 208–220 (2006).
McGregor, M. J. & Muskal, S. M. Pharmacophore fingerprinting. 1. Application to QSAR and focused library design. J. Chem. Inf. Model. 39, 569–574 (1999).
Hall, L. H. & Kier, L. B. Electrotopological state indices for atom types: a novel combination of electronic, topological, and valence state information. J. Chem. Inf. Comput. Sci. 35, 1039–1045 (1995).
Sha, Y., Phan, J. H. & Wang, M. D. Effect of low-expression gene filtering on detection of differentially expressed genes in RNA-seq data. In 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society(EMBS) 6461–6464 (Institute of Electrical and Electronics Engineers, 2015).
Jonker, R. & Volgenant, A. A shortest augmenting path algorithm for dense and sparse linear assignment problems. Computing 38, 325–340 (1987).
Chen, Y. et al. Dual path networks. Adv. Neural Inf. Process. Syst. 30, 4467–4475 (2017).
Skinnider, M. A., Dejong, C. A., Franczak, B. C., McNicholas, P. D. & Magarvey, N. A. Comparative analysis of chemical similarity methods for modular natural products with a hypothetical structure enumeration algorithm. J. Cheminform. 9, 46 (2017).
Škuta, C. et al. QSAR-derived affinity fingerprints (part 1): fingerprint construction and modeling performance for similarity searching, bioactivity classification and scaffold hopping. J. Cheminform. 12, 39 (2020).
Fernández-de Gortari, E., García-Jacas, C. R., Martinez-Mayorga, K. & Medina-Franco, J. L. Database fingerprint (DFP): an approach to represent molecular databases. J. Cheminform. 9, 9 (2017).
Sato, T., Honma, T. & Yokoyama, S. Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J. Chem. Inf. Model. 50, 170–185 (2010).
Smusz, S., Kurczab, R. & Bojarski, A. J. The influence of the inactives subset generation on the performance of machine learning methods. J. Cheminform. 5, 17 (2013).
Czarnecki, W. M., Podlewska, S. & Bojarski, A. J. Robust optimization of SVM hyperparameters in the classification of bioactive compounds. J. Cheminform. 7, 38 (2015).
Askjaer, S. & Langgård, M. Combining pharmacophore fingerprints and PLS-discriminant analysis for virtual screening and SAR elucidation. J. Chem. Inf. Model. 48, 476–488 (2008).
Venkatraman, V., Pérez-Nueno, V. I., Mavridis, L. & Ritchie, D. W. Comprehensive comparison of ligand-based virtual screening tools against the DUD data set reveals limitations of current 3D methods. J. Chem. Inf. Model. 50, 2079–2093 (2010).
Szegedy, C. et al. Going deeper with convolutions. In IEEE Conference on Computer Vision and Pattern Recognition 1–9 (CVPR, 2015).
Mayr, A. et al. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem. Sci. 9, 5441–5451 (2018).
Ramsundar, B., Eastman, P., Walters, P. & Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More (O’Reilly Media, 2019).
Shen, W. X. et al. ChemBench: the molecule benchmarks and MolMapNet datasets. Zenodo https://doi.org/10.5281/zenodo.4054866 (2020).
Shen, W. X. et al. The molmap package. Zenodo https://doi.org/10.5281/zenodo.4056290 (2020).
Acknowledgements
We appreciate the financial support from the National Key R&D Program of China, Synthetic Biology Research (2019YFA0905900), Shenzhen Municipal Government grants (no. 2019156, JCYJ20170413113448742 and no. 201901), Department of Science and Technology of Guangdong Province (no. 2017B030314083) and Singapore Academic Funds R-148-000-273-114. We thank P. J. Feng for his help on providing CYP450 data. We also thank S. Liang and X. Zeng for their help in data analysis and evaluations.
Author information
Authors and Affiliations
Contributions
Y.Z.C and W.X.S. designed the study and wrote the manuscript. W.X.S. and X.Z. performed the experiments and data analysis. Y.Y.J. and Y.Z. provided the experimental platform, F.Z., Y.T., C.Q. and Y.W. provided evaluation and suggestions. All authors contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The performance of the GCN/GAT models and the MolMapNet-OOTB model on 6 single-task benchmark datasets under 10 different splits.
MolMapNet-OOTB model is compared to the D-MPNN and AttentiveFP models, the 10 different random seeds 2, 16, 32, 64, 128, 256, 512, 1024, 2048, and 4096 were used for splitting the training set (0.8), validation set (0.1) and test set (0.1). a, 3 regression tasks: Malaria, ESOL, FreeSolv under random split. b, 3 classification tasks (BACE, BBBP, and HIV) under the scaffold-split.
Extended Data Fig. 2 The performance of the GCN/GAT models and the MolMapNet-OOTB model on 6 multi-task benchmarks under 10 different splits.
MolMapNet-OOTB model is compared to the D-MPNN and AttentiveFP models, the 10 different random seeds 2, 16, 32, 64, 128, 256, 512, 1024, 2048, and 4096 were used to split the training set (0.8), validation set (0.1) and test set (0.1). a, 3 classification tasks (Tox21, ToxCast, and SIDER) under random split. b, 3 high-data classification tasks (MUV, PCBA, and ChEMBL) under random split.
Extended Data Fig. 3 The performance of single-path MolMapNet-D, MolMapNet-F and dual-path MolMapNet-B models on 11 benchmarks.
a, 5 regression benchmark datasets of metric RMSE (ESOL, FreeSolv, Lipop, PDBbind-F, Malaria). b, 6 classification benchmark datasets of metric ROC_AUC (BACE, BBBP, HIV, ClinTox, SIDER, Tox21,). These benchmarks are split into training, validation and test set by using both MoleculeNet data-splits (labeled as, for example, ESOL-M) and AttentiveFP data-splits (labeled as, for example, ESOL-A). Note: the error bars represent standard error of the mean.
Extended Data Fig. 4 The performance of kNN and MolMapNet-F on the 5 classification tasks.
The 5 classification tasks are under the MoleculeNet data splits, both kNN and MolMapNet-F are based on three sets of fingerprints: PubChemFP, MACCSFP, and PharmacoErGFP (PubFP-MACFP-ErGFP), the error bars represent standard error of the mean.
Extended Data Fig. 5 Optimization of MolMapNet feature-generation parameters n_neighbors and min_dist using grid-search strategy.
The parameters n_neighbors and min_dist are in the range of 10~105 and 0~1 respectively, the three datasets ESOL, BACE, and Tox21 are split by the MoleculeNet data-splits method. a, optimization of MolMapNet-D model on the ESOL dataset, the performance was evaluated by RMSE of the validation set. b, optimization of MolMapNet-F model on the BACE dataset, the performance was evaluated by ROC-AUC of the validation set. c, optimization of MolMapNet-B model on the Tox21 dataset, the performance was evaluated by ROC-AUC of the validation set.
Extended Data Fig. 6 The TMAP visualization of the BACE training, validation, test and the novel ChEMBL set represented by the 1024-bit Morgan fingerprint(r=2).
a, the similarity distribution of the four sets in different color by TMAP36: the train_data, valid_data and test_data are the training (646 high potency inhibitors, 564 low potency inhibitors), validation (77 high potency inhibitors, 74 low potency inhibitors), and testing (50 high potency inhibitors, 102 low potency inhibitors) set split from the BACE benchmark dataset using the scaffold-split method, the novel_data is the novel ChEMBL set (216 BACE high potency inhibitors, 179 low potency inhibitors from the ChEMBL database). b, the distribution of the compounds with respect to activity type (BACE high potency inhibitors in green and low potency inhibitors in blue color), the interactive visualization is provided at: http://bidd.group/molmap/BACE/BACE.html.
Extended Data Fig. 7 The PCA of the latent features of the global max pooling (GMP) layer of the MolMapNet-D solubility model and the MolMapNet-F BACE inhibitor model.
a, the MolMapNet-D solubility model. b, the MolMapNet-F BACE inhibitor model. The MolMapNet-D solubility model was trained on the ESOL benchmark dataset using the AttentiveFP data-split. The MolMapNet-F BACE benchmark model was trained on the BACE dataset using the AttentiveFP data-split (scaffold split).
Extended Data Fig. 8 The important input-features of the MolMapNet-D solubility model trained on the ESOL dataset using the AttentiveFP data-split.
a, the feature importance score of the important features for the ESOL training vs. the test set. b, the attention map (the heatmap of the feature importance value). Features of higher positive scores are of higher importance. Features of negative score adversely affect model performance. The top important features are Estate, Charge, Matrix and several other descriptors concentrated in the specific red, orange, and bright green regions in b.
Extended Data Fig. 9 The important input-features of the BACE inhibitor classification MolMapNet-F model.
a, the feature importance score of the important features for the BACE training vs. test set (the Pearson correlation coefficient between the two sets is 0.887). and the model attention map (the heatmap of the feature importance value, the smarts patterns of the fingerprint features in the six annotated groups are provided in Supplementary Table 8). b, the three groups of the important fingerprints. c, the proportion of the top 50 important features and the bottom 50 features in the BACE high potency and low potency inhibitors.
Extended Data Fig. 10 The average importance of the atoms and bonds of the BACE inhibitors of two molecular scaffolds in the BACE benchmark dataset.
The two molecular scaffolds of BACE inhibitors are 2-aminoquinoline44 and 2-aminobenzimidazole45, the atoms and bonds of each inhibitor are color-highlighted based on the presence of top50 important features (green color indicates higher average importance, red color lower importance), and their bioactivity in pIC50 values are provided. Compounds with higher portions of the important features (green) tend to have higher activity values. The substructures in the dotted circles are consistent with literature-reported structure-activity relationships of BACE inhibitors in previous study44.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1–9 and Methods 1–5.
Source data
Source Data Fig. 1
The embedding coordinates data and grid assignment coordinates of the feature points.
Source Data Fig. 2
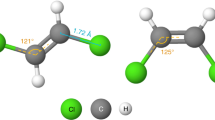
Source data of aspirin and analogue feature maps.
Source Data Extended Data Fig. 1
Raw data of the boxplot and mean values.
Source Data Extended Data Fig. 2
Raw data of the boxplot and mean values.
Source Data Extended Data Fig. 3
The MolMapNet OOTB and MolMapNet OPT model performance data.
Source Data Extended Data Fig. 4
The MolMapNet-F and kNN model performance data.
Source Data Extended Data Fig. 5
The grid search results data.
Source Data Extended Data Fig. 6
The coordinates of the TMAP results, also includes the relevant data: the novel and common BACE data from ChEMBL and the collected clinical BACE drugs.
Source Data Extended Data Fig. 7
The PCA results data of the outputs of GMP layers.
Source Data Extended Data Fig. 8
The feature importance source data of the figure.
Source Data Extended Data Fig. 9
The feature importance data and the statistical proportion of these feature points.
Rights and permissions
About this article
Cite this article
Shen, W.X., Zeng, X., Zhu, F. et al. Out-of-the-box deep learning prediction of pharmaceutical properties by broadly learned knowledge-based molecular representations. Nat Mach Intell 3, 334–343 (2021). https://doi.org/10.1038/s42256-021-00301-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00301-6