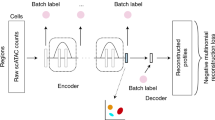
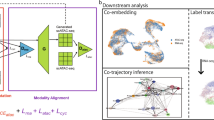
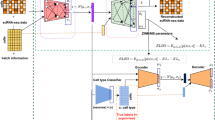
Abstract
Recent advances in single-cell technologies, including single-cell ATAC-seq (scATAC-seq), have enabled large-scale profiling of the chromatin accessibility landscape at the single-cell level. However, the characteristics of scATAC-seq data, including high sparsity and high dimensionality, have greatly complicated the computational analysis. Here, we propose scDEC, a computational tool for scATAC-seq analysis with deep generative neural networks. scDEC is built on a pair of generative adversarial networks, and is capable of simultaneously learning the latent representation and inferring cell labels. In a series of experiments, scDEC demonstrates superior performance over other tools in scATAC-seq analysis across multiple datasets and experimental settings. In downstream applications, we demonstrate that the generative power of scDEC helps to infer the trajectory and intermediate state of cells during differentiation and the latent features learned by scDEC can potentially reveal both biological cell types and within-cell-type variations. We also show that it is possible to extend scDEC for the integrative analysis of multi-modal single cell data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The InSilico dataset was collected from the GEO database with accession number GSE65360. The mouse Forebrain dataset was downloaded from the GEO database with accession number GSE100033. The Splenocyte dataset can be accessed at ArrayExpress database with accession number E-MTAB-6714. The All blood dataset can be accessed at the GEO database with accession number GSE96772. The mouse atlas data are available at http://atlas.gs.washington.edu/mouse-atac. The human PBMCs dataset used in multi-modal single cell analysis was downloaded from 10x Genomics (https://support.10xgenomics.com/single-cell-multiome-atac-gex) with entry ‘pbmc_granulocyte_sorted_10k’. The preprocessed scATAC-seq data used as input for scDEC model in this study can be downloaded from https://doi.org/10.5281/zenodo.397785856.
Code availability
scDEC is open-source software based on the TensorFlow library57, which is available on Github (https://github.com/kimmo1019/scDEC) and Zenodo (https://doi.org/10.5281/zenodo.4560834)58. A CodeOcean capsule with several example datasets is available at https://codeocean.com/capsule/0746056/tree/v159. The pretrained models on both benchmark single-cell datasets and 10x Genomics PBMCs multi-modal single-cell dataset were provided.
References
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Cusanovich, D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Chen, H. et al. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 20, 241 (2019).
Zamanighomi, M. et al. Unsupervised clustering and epigenetic classification of single cells. Nat. Commun. 9, 2410 (2018).
González-Blas, C. B. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e1318 (2018).
Baker, S. M., Rogerson, C., Hayes, A., Sharrocks, A. D. & Rattray, M. Classifying cells with Scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Res. 47, e10 (2019).
Fang, R. et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021).
Goodfellow, I. et al. Generative adversarial nets. In Proceedings of Advances in Neural Information Processing Systems (NeurIPS) 2672–2680 (NIPS, 2014).
Kingma, D. P. & Welling, M. Auto-encoding variational bayes. In Proceedings of International Conference on Learning Representations (ICLR, 2014).
Liu, Q., Lv, H. & Jiang, R. hicGAN infers super resolution Hi-C data with generative adversarial networks. Bioinformatics 35, i99–i107 (2019).
Xiong, L. et al. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat. Commun. 10, 4576 (2019).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision 2223–2232 (ICCV, 2017).
Liu, Q., Xu, J., Jiang, R. & Wong, W. H. Density estimation using deep generative neural networks. Proc. Natl Acad. Sci. USA 118, e2101344118 (2021).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection. J. Open Source Software 3, 861 (2018).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Preissl, S. et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 21, 432–439 (2018).
Chen, X., Miragaia, R. J., Natarajan, K. N. & Teichmann, S. A. A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 9, 5345 (2018).
Buenrostro, J. D. et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548 (2018).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Mathelier, A. et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–115 (2016).
Shaltouki, A., Peng, J., Liu, Q., Rao, M. S. & Zeng, X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells 31, 941–952 (2013).
Bayam, E. et al. Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation. BMC Genomics 16, 681 (2015).
Owa, T. et al. Meis1 coordinates cerebellar granule cell development by regulating Pax6 transcription, BMP signaling and Atoh1 degradation. J. Neurosci. 38, 1277–1294 (2018).
Hallonet, M., Hollemann, T., Pieler, T. & Gruss, P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 13, 3106–3114 (1999).
Cesari, F. et al. Mice deficient for the Ets transcription factor Elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol. Cell. Biol. 24, 294–305 (2004).
Stolt, C. C. et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 (2003).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Iwasaki, H. & Akashi, K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity 26, 726–740 (2007).
Gilmour, J. et al. A crucial role for the ubiquitously expressed transcription factor Sp1 at early stages of hematopoietic specification. Development 141, 2391–2401 (2014).
Anderson, K. C. et al. Expression of human B cell-associated antigens on leukemias and lymphomas: a model of human B cell differentiation. Blood 63, 1424–1433 (1984).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Argelaguet, R. et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 21, 111 (2020).
Jin, S., Zhang, L. & Nie, Q. scAI: an unsupervised approach for the integrative analysis of parallel single-cell transcriptomic and epigenomic profiles. Genome Biol. 21, 25 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Teller, V. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. Comput. Linguist. 26, 638–641 (2000).
Chowdhury, G. G. Introduction to Modern Information Retrieval (Facet, 2010).
Halko, N., Martinsson, P.-G. & Tropp, J. A. Finding structure with randomness: probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev. 53, 217–288 (2011).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Gulrajani, I., Ahmed, F., Arjovsky, M., Dumoulin, V. & Courville, A. C. Improved training of Wasserstein GANs. In Proceedings of Advances in Neural Information Processing Systems 5767–5777 (NIPS, 2017).
Yi, Z., Zhang, H., Tan, P. & Gong, M. Dualgan: Unsupervised dual learning for image-to-image translation. In Proceedings of the IEEE International Conference on Computer Vision 2849–2857 (ICCV, 2017).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In Proceedings of International Conference on Learning Representations (ICLR, 2014).
Mukherjee, S., Asnani, H., Lin, E. & Kannan, S. In Proceedings of the AAAI Conference on Artificial Intelligence Vol. 33, 4610–4617 (AAAI, 2019).
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning 448–456 (ICML, 2015).
Strehl, A. & Ghosh, J. Cluster ensembles—a knowledge reuse framework for combining multiple partitions. J. Mach. Learn. Res. 3, 583–617 (2002).
Hubert, L. & Arabie, P. Comparing partitions. J. Classification 2, 193–218 (1985).
Rosenberg, A. & Hirschberg, J. V-measure: A conditional entropy-based external cluster evaluation measure. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning 410–420 (EMNLP-CoNLL, 2007).
Rand, W. M. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66, 846–850 (1971).
Tibshirani, R., Walther, G. & Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. B 63, 411–423 (2001).
Mann, H. B. & Whitney, D. R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60 (1947).
Liu, Q. et al. scDEC: data for simultaneous deep generative modeling and clustering of single cell genomic data. Zenodo https://doi.org/10.5281/zenodo.3984189 (2020).
Abadi, M. et al. Tensorflow: a system for large-scale machine learning. In Proceedings of 12th USENIX Symposium on Operating Systems Design and Implementation 265–283 (OSDI, 2016).
Liu, Q. et al. scDEC: code for simultaneous deep generative modeling and clustering of single cell genomic data. Zenodo https://doi.org/10.5281/zenodo.4560834 (2021).
Liu, Q. et al. scDEC: simultaneous deep generative modeling and clustering of single cell genomic data. CodeOcean https://doi.org/10.24433/CO.3347162.v1 (2020).
Acknowledgements
This work was supported by NIH grants R01 HG010359 (W.H.W.) and P50 HG007735 (W.H.W.). This work was also supported by the National Key Research and Development Program of China no. 2018YFC0910404 (R.J.), the National Natural Science Foundation of China nos 61873141 (R.J.), 61721003 (R.J.) and 61573207 (R.J.).
Author information
Authors and Affiliations
Contributions
W.H.W., R.J. and Q.L. conceived the study. Q.L. designed and implemented scDEC. Q.L., S.C. and W.H.W. performed the data analysis. Q.L. and W.H.W. interpreted the results. Q.L., R.J. and W.H.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1–6.
Rights and permissions
About this article
Cite this article
Liu, Q., Chen, S., Jiang, R. et al. Simultaneous deep generative modelling and clustering of single-cell genomic data. Nat Mach Intell 3, 536–544 (2021). https://doi.org/10.1038/s42256-021-00333-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00333-y
This article is cited by
-
DANCE: a deep learning library and benchmark platform for single-cell analysis
Genome Biology (2024)
-
scCASE: accurate and interpretable enhancement for single-cell chromatin accessibility sequencing data
Nature Communications (2024)
-
LSH-GAN enables in-silico generation of cells for small sample high dimensional scRNA-seq data
Communications Biology (2022)
-
Simultaneous dimensionality reduction and integration for single-cell ATAC-seq data using deep learning
Nature Machine Intelligence (2022)
-
scBasset: sequence-based modeling of single-cell ATAC-seq using convolutional neural networks
Nature Methods (2022)