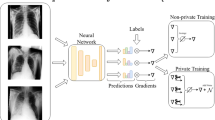
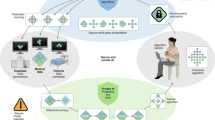
Abstract
Using large, multi-national datasets for high-performance medical imaging AI systems requires innovation in privacy-preserving machine learning so models can train on sensitive data without requiring data transfer. Here we present PriMIA (Privacy-preserving Medical Image Analysis), a free, open-source software framework for differentially private, securely aggregated federated learning and encrypted inference on medical imaging data. We test PriMIA using a real-life case study in which an expert-level deep convolutional neural network classifies paediatric chest X-rays; the resulting model’s classification performance is on par with locally, non-securely trained models. We theoretically and empirically evaluate our framework’s performance and privacy guarantees, and demonstrate that the protections provided prevent the reconstruction of usable data by a gradient-based model inversion attack. Finally, we successfully employ the trained model in an end-to-end encrypted remote inference scenario using secure multi-party computation to prevent the disclosure of the data and the model.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The paediatric pneumonia dataset is publicly available from Mendeley Data at https://doi.org/10.17632/rscbjbr9sj.3. The MedNIST dataset was assembled by B. J. Erickson (Department of Radiology, Mayo Clinic) and is available at https://github.com/Project-MONAI/MONAI/. The MSD Liver Segmentation Dataset is available at http://medicaldecathlon.com. test sets 1 and 2 contain confidential patient information and cannot be shared publicly. Source data are provided with this paper.
Code availability
The current version of the PriMIA source code is publicly available at https://github.com/gkaissis/PriMIA and permanently archived at https://doi.org/10.5281/zenodo.454559918. PriMIA includes source code from PySyft (https://github.com/OpenMined/PySyft), PyGrid (https://github.com/OpenMined/PyGrid) and Opacus (https://github.com/pytorch/opacus) re-used under open-source licence terms.
References
McKinney, S. M. et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020).
Ardila, D. et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 25, 954–961 (2019).
Patent Index 2019: Spotlight on digital technologies. European Patent Office https://www.epo.org/about-us/annual-reports-statistics/statistics/2019.html (accessed 10 March 2021).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Sheller, M. J. et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 10, 12598 (2020).
Schwarz, C. G. et al. Identification of anonymous MRI research participants with face-recognition software. N. Engl. J. Med. 381, 1684–1686 (2019).
Narayanan, A. & Shmatikov, V. Robust de-anonymization of large sparse datasets. In 2008 IEEE Symposium on Security and Privacy (sp 2008) 111–125 (IEEE, 2008).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Banerjee, S., Hemphill, T. & Longstreet, P. Wearable devices and healthcare: data sharing and privacy. Inf. Soc. 34, 49–57 (2018).
Raisaro, J. L. et al. SCOR: a secure international informatics infrastructure to investigate COVID-19. J. Am. Med. Inform. Assoc. 27, 1721–1726 (2020).
Vaid, A. et al. Federated learning of electronic health records to improve mortality prediction in hospitalized patients with COVID-19: machine learning approach. JMIR Med. Inform. 9, e24207 (2021).
Roth, H. R. et al. Federated learning for breast density classification: a real-world implementation. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning 181–191 (Springer, 2020).
Fredrikson, M., Jha, S. & Ristenpart, T. Model inversion attacks that exploit confidence information and basic countermeasures. In Proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security (CCS 2015) (ACM Press, 2015).
Wang, Z. et al. Beyond inferring class representatives: user-level privacy leakage from federated learning. In IEEE INFOCOM 2019-IEEE Conference on Computer Communications 2512–2520 (IEEE, 2019).
La, H. J., Kim, M. K. & Kim, S. D. A personal healthcare system with inference-as-a-service. In 2015 IEEE International Conference on Services Computing 249–255 (IEEE, 2015).
Kaissis, G. A., Makowski, M. R., Rückert, D. & Braren, R. F. Secure, privacy-preserving and federated machine learning in medical imaging. Nat. Mach. Intell. 2, 305–311 (2020).
Ryffel, T. et al. A generic framework for privacy preserving deep learning. Preprint at https://arxiv.org/abs/1811.04017 (2018).
Kaissis, G. & Ziller, A. PriMIA version 2021.02 https://doi.org/10.5281/zenodo.4545599 (2021).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 770–778 (2016).
Kermany, D. S. et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172, 1122–1131 (2018).
Gupta, G. R. Tackling pneumonia and diarrhoea: the deadliest diseases for the world’s poorest children. Lancet 379, 2123–2124 (2012).
Evans, D., Kolesnikov, V. & Rosulek, M. A pragmatic introduction to secure multi-party computation. Found. Trends Privacy Secur. 2, 70–246 (2018).
Abadi, M. et al. Deep learning with differential privacy. In Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security (ACM, 2016).
Mironov, I., Talwar, K. & Zhang, L. Rényi differential privacy of the sampled gaussian mechanism. Preprint at https://arxiv.org/abs/1908.10530 (2019).
Damgård I., Pastro V., Smart N. & Zakarias S. Multiparty computation from somewhat homomorphic encryption. In Advances in Cryptology – CRYPTO 2012 (eds. Safavi-Naini, R. & Canetti R.) (Springer, 2012).
Ryffel, T., Pointcheval, D. & Bach, F. ARIANN: low-interaction privacy-preserving deep learning via function secret sharing. Preprint at https://arxiv.org/abs/2006.04593 (2020).
Matthews, B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta Protein Struct. 405, 442–451 (1975).
Boyle, E., Gilboa, N. & Ishai, Y. Function secret sharing. In Annual International Conference on the Theory and Applications of Cryptographic Techniques 337–367 (Springer, 2015).
Wagh, S., Gupta, D., & Chandran, N. Securenn: 3-party secure computation for neural network training. In Proc. Privacy Enhancing Technologies 26–49 (Sciendo, 2019).
Carlini, N., Liu, C., Erlingsson, Ú., Kos, J. & Song, D. The secret sharer: evaluating and testing unintended memorization in neural networks. In 28th {USENIX} Security Symposium ({USENIX} Security 19) 267–284 (2019).
Zhao, B., Mopuri, K. R. & Bilen, H. iDLG: improved deep leakage from gradients. Preprint at https://arxiv.org/abs/2001.02610 (2020).
Geiping, J., Bauermeister, H., Dröge, H. & Moeller, M. Inverting gradients. How easy is it to break privacy in federated learning? In Advances in Neural Information Processing Systems 16937–16947 (NeurIPS, 2020).
Bluemke, D. A. et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the radiology editorial board. Radiology 294, 487–489 (2020).
Wu, B. et al. P3SGD: patient privacy preserving SGD for regularizing deep CNNs in pathological image classification P3SGD. In Proc. Conference on Computer Vision and Pattern Recognition 2099–2108 (CVPR, 2019).
Reddi, S. et al. Adaptive federated optimization. Preprint at https://arxiv.org/abs/2003.00295 (2020).
Fu, Y., Wang, H., Xu, K., Mi, H. & Wang, Y. Mixup based privacy preserving mixed collaboration learning. In 2019 IEEE International Conference on Service-Oriented System Engineering (SOSE) 275–2755 (IEEE, 2019).
McMahan, B., Moore, E., Ramage, D., Hampson, S. & y Arcas, B. A. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics 1273–1282 (PMLR, 2017).
Bergstra, J., Bardenet, R., Bengio, Y. & Kégl, B. Algorithms for hyper-parameter optimization. In Proceedings of the 24th International Conference on Neural Information Processing Systems 2546–2554 (Curran Associates, 2011).
Parks, C. L. & Monson, K. L. Automated facial recognition of computed tomography-derived facial images: patient privacy implications. J. Digital Imaging 30, 204–214 (2016).
Qaisar Ahmad Al Badawi, A. et al. Towards the AlexNet moment for homomorphic encryption: HCNN, the first homomorphic CNN on encrypted data with GPUs. In IEEE Transactions on Emerging Topics in Computing (IEEE, 2020).
Wagh, S. et al. Falcon: honest-majority maliciously secure framework for private deep learning. In Proc. Privacy Enhancing Technologies 188–208 (Sciendo, 2021).
Silva, S., Altmann, A., Gutman, B. & Lorenzi, M. Fed-BioMed: a general open-source frontend framework for federated learning in healthcare. In Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning (eds Albarqouni, S. et al.) 201–210 (Springer, 2020).
Sheller, M. J., Reina, G. A., Edwards, B., Martin, J. & Bakas, S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries 92–104 (Springer, 2019).
Li, W. et al. Privacy-preserving federated brain tumour segmentation. In International Workshop on Machine Learning in Medical Imaging 133–141 (Springer, 2019).
Lu, M. Y. et al. Federated learning for computational pathology on gigapixel whole slide images. Preprint at https://arxiv.org/abs/2009.10190 (2020).
Li, X. et al. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal. 65, 101765 (2020).
Kairouz, P. & McMahan, H. B. Advances and Open Problems in Federated Learning (Now, 2021).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. In Conference on Computer Vision and Pattern Recognition (CVPR09) (2009).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision 1026–1034 (2015).
Buslaev, A. et al. Albumentations: fast and flexible image augmentations. Information 11, 125 (2020).
Huang, L., Zhang, C. & Zhang, H. Self-adaptive training: beyond empirical risk minimization. In Advances in Neural Information Processing Systems Vol. 33 (NeurIPS, 2020).
Kingma, P. & Ba, J. Adam: a method for stochastic optimization. In Proc. International Conference on Learning Representations (ICLR, 2015).
Rieke, N. et al. The future of digital health with federated learning. npj Digital Med. 3, 119 (2020).
Bonawitz, K. et al. Practical secure aggregation for federated learning on user-held data. In NIPS Workshop on Private Multi-Party Machine Learning (NIPS, 2016).
Wang, S. et al. Adaptive federated learning in resource constrained edge computing systems. IEEE J. Sel. Areas Commun. 37, 1205–1221 (2019).
Kirkpatrick, J. et al. Overcoming catastrophic forgetting in neural networks. Proc. Natl Acad. Sci. USA 114, 3521–3526 (2017).
Boyle, E., Gilboa, N. & Ishai, Y. Function secret sharing: improvements and extensions. In Proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security 1292–1303 (2016).
Boyle, E., Gilboa, N. & Ishai, Y. Secure computation with preprocessing via function secret sharing. In Theory of Cryptography Conference 341–371 (Springer, 2019).
Shokri, R., Stronati, M., Song, C. & Shmatikov, V. Membership inference attacks against machine learning models. In 2017 IEEE Symposium on Security and Privacy (SP) 3–18 (IEEE, 2017).
He, Z., Zhang, T. & Lee, R. B. Model inversion attacks against collaborative inference. In Proceedings of the 35th Annual Computer Security Applications Conference (ACM, 2019).
Chicco, D. & Jurman, G. The advantages of the matthews correlation coefficient (mcc) over f1 score and accuracy in binary classification evaluation. BMC Genomics 21, 6 (2020).
Zhu, L., Liu, Z. & Han, S. Deep leakage from gradients. In Advances in Neural Information Processing Systems 14774–14784 (2019).
Wang, Y. et al. SAPAG: a self-adaptive privacy attack from gradients. Preprint at https://arxiv.org/abs/2009.06228 (2020).
Oh, H. & Lee, Y. Exploring image reconstruction attack in deep learning computation offloading. In The 3rd International Workshop on Deep Learning for Mobile Systems and Applications: EMDL ’19 (ACM, 2019).
Gao, W. et al. Privacy-preserving collaborative learning with automatic transformation search. In Proc. Conference on Computer Vision and Pattern Recognition (CVPR, 2021).
Yanchun, L. & Nanfeng, X. Generative adversarial networks based on denoising and reconstruction regularization. In 2019 IEEE 21st International Conference on High Performance Computing and Communications IEEE 17th International Conference on Smart City IEEE 5th International Conference on Data Science and Systems (HPCC/SmartCity/DSS) (IEEE, 2019).
Kermany, D., Zhang, K. & Goldbaum, M. Large dataset of labeled optical coherence tomography (OCT) and chest X-ray images. Mendeley Data https://doi.org/10.17632/rscbjbr9sj.3 (2018).
Acknowledgements
We acknowledge funding from the following sources, funders played no role in the design of the study, the preparation of the manuscript or the decision to publish. The Technical University of Munich, School of Medicine Clinician Scientist Programme (KKF), project reference H14 (G.K.). German Research Foundation, SPP2177/1, German Cancer Consortium (DKTK) and TUM Foundation, Technical University of Munich (R.B. and G.K.). European Community Seventh Framework Programme (FP7/2007-2013 grant no. 339563 – CryptoCloud) and FUI ANBLIC Project (T.R.). UK Research and Innovation London Medical Imaging & Artificial Intelligence Centre for Value Based Healthcare (D.R. and G.K.). Technical University Munich/ Imperial College London Joint Academy of Doctoral Studies (D.U.). We thank B. Farkas for creating Figures 1 and 2 as well as the PriMIA logo; P. Cason and H. Emanuel for assisting with PyGrid debugging, M. Lau for his input, D. Testuggine for his input on differentially private gradient descent, M. Jay for helping with PySyft debugging, N. Remerscheid for his work on the liver segmentation case study, the PySyft and PyGrid development teams for their foundational work and the OpenMined community for their scientific input, contributions and discussion.
Author information
Authors and Affiliations
Contributions
G.K. conceived and coordinated the project, evaluated the chest radiography data, helped with PriMIA programming and wrote the paper. A.Z. conceived and developed PriMIA, oversaw PriMIA development, trained the models, performed data analysis and wrote the paper. J.P.-P. helped with project management, oversaw the security and cryptography aspects of PriMIA, supervised model inversion attacks and helped write the paper. T.R. designed and developed PySyft and PyGrid, designed and implemented the AriaNN FSS protocol, helped with PriMIA programming and performed inference latency assessment. D.U. performed the model inversion attacks and helped write the paper. A.T. conceived the OpenMined project, designed and developed PySyft and PyGrid, provided project guidance and assistance and helped with PriMIA development. I.D.L.C.J. developed PyGrid and helped with PriMIA programming. J.M. provided project guidance and prototype code. F.J. performed data curation and helped with data analysis on the chest radiographs. M.-M.S. performed data curation and evaluated the chest radiography data. A.S. and M.M. provided project management, support and guidance. D.R. provided oversight, project management, support, guidance and scientific input, and helped write the paper. R.B. provided oversight, project management, support and guidance, helped with data procurement, provided scientific input and helped write the paper. All authors proof-read and accepted the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Haixu Tang, Holger Roth and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Tables 1 and 2 and methods/discussion.
Source data
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 1
Figure Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Rights and permissions
About this article
Cite this article
Kaissis, G., Ziller, A., Passerat-Palmbach, J. et al. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat Mach Intell 3, 473–484 (2021). https://doi.org/10.1038/s42256-021-00337-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00337-8
This article is cited by
-
Real world federated learning with a knowledge distilled transformer for cardiac CT imaging
npj Digital Medicine (2025)
-
Private pathological assessment via machine learning and homomorphic encryption
BioData Mining (2024)
-
Fog-based deep learning framework for real-time pandemic screening in smart cities from multi-site tomographies
BMC Medical Imaging (2024)
-
Preserving fairness and diagnostic accuracy in private large-scale AI models for medical imaging
Communications Medicine (2024)
-
Generative models improve fairness of medical classifiers under distribution shifts
Nature Medicine (2024)