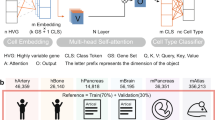
Abstract
Annotating cell types on the basis of single-cell RNA-seq data is a prerequisite for research on disease progress and tumour microenvironments. Here we show that existing annotation methods typically suffer from a lack of curated marker gene lists, improper handling of batch effects and difficulty in leveraging the latent gene–gene interaction information, impairing their generalization and robustness. We developed a pretrained deep neural network-based model, single-cell bidirectional encoder representations from transformers (scBERT), to overcome the challenges. Following BERT’s approach to pretraining and fine-tuning, scBERT attains a general understanding of gene–gene interactions by being pretrained on huge amounts of unlabelled scRNA-seq data; it is then transferred to the cell type annotation task of unseen and user-specific scRNA-seq data for supervised fine-tuning. Extensive and rigorous benchmark studies validated the superior performance of scBERT on cell type annotation, novel cell type discovery, robustness to batch effects and model interpretability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data used in this study are publicly available and the usages are fully illustrated in the Methods. The published Panglao dataset was downloaded from https://panglaodb.se/. The published Zheng68k dataset was downloaded from the ‘Fresh 68K PBMCs’ section at https://support.10xgenomics.com/single-cell-gene-expression/datasets (SRP073767)34. The published pancreatic datasets were downloaded from github at https://hemberg-lab.github.io/scRNA.seq.datasets/ (Baron: GSE84133, Muraro: GSE85241, Segerstolpe: E-MTAB-5061, Xin: GSE81608)35,36,37,38. The MacParland dataset was downloaded from https://www.ncbi.nlm.nih.gov/geo/ (GSE115469)50. The heart datasets were downloaded from https://data.humancellatlas.org/explore/projects/ad98d3cd-26fb-4ee3-99c9-8a2ab085e737 and https://singlecell.broadinstitute.org/single_cell/study/SCP498/transcriptional-and-cellular-diversity-of-the-human-heart (refs. 51,52). The lung dataset for COVID-19 study was downloaded from https://doi.org/10.6084/m9.figshare.11981034.v1 (ref. 53). The adult Human Cell Atlas of 15 major organs dataset was downloaded from https://www.ncbi.nlm.nih.gov/geo/ (GSE159929)54. Source Data are provided with this paper.
Code availability
The source code of the pre-processing, scBERT modelling and fine-tuning processes are freely available on Github (https://github.com/TencentAILabHealthcare/scBERT) and Zenodo (https://doi.org/10.5281/zenodo.6572672)60 with detailed instructions. The source code for the other comparison methods are publicly available (see Supplementary Table 2).
References
Plass, M. et al. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, aaq1723 (2018).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Zhao, X., Wu, S., Fang, N., Sun, X. & Fan, J. Evaluation of single-cell classifiers for single-cell RNA sequencing data sets. Briefings Bioinform. 21, 1581–1595 (2020).
Pasquini, G., Rojo Arias, J. E., Schäfer, P. & Busskamp, V. Automated methods for cell type annotation on scRNA-seq data. Comput. Struct. Biotechnol. J.19, 961–969 (2021).
Cao, Y., Wang, X. & Peng, G. SCSA: a cell type annotation tool for single-cell RNA-seq data. Front. Genet. 0, 490 (2020).
Huang, Q., Liu, Y., Du, Y. & Garmire, L. X. Evaluation of cell type annotation R packages on single-cell RNA-seq data. Genomics Proteomics Bioinform. 19, 267–281 (2020).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, aau5324 (2018).
Zhang, Z. et al. SCINA: a semi-supervised subtyping algorithm of single cells and bulk samples. Genes 10, 531 (2019).
Pliner, H. A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 16, 983–986 (2019).
Grabski, I. N. & Irizarry, R. A. A probabilistic gene expression barcode for annotation of cell types from single-cell RNA-seq data. Biostatistics. https://doi.org/10.1093/biostatistics/kxac021 (2022).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 1–32 (2020).
Serra, A., Coretto, P., Fratello, M. & Tagliaferri, R. Robust and sparse correlation matrix estimation for the analysis of high-dimensional genomics data. Bioinformatics 34, 625–634 (2018).
Ma, F. & Pellegrini, M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics 36, 533–538 (2020).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 1–17 (2019).
Cao, Z.-J., Wei, L., Lu, S., Yang, D.-C. & Gao, G. Searching large-scale scRNA-seq databases via unbiased cell embedding with Cell BLAST. Nature Commun. 11, 1–13 (2020).
Xie, P. et al. SuperCT: a supervised-learning framework for enhanced characterization of single-cell transcriptomic profiles. Nucleic Acids Res. 47, e48–e48 (2019).
Li, C. et al. SciBet as a portable and fast single cell type identifier. Nat. Commun. 11, 1–8 (2020).
Qiu, P. Embracing the dropouts in single-cell RNA-seq analysis. Nat. Commun. 11, 1–9 (2020).
Wang, T. et al. MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat. Commun. 12, 1–13 (2021).
Wang, T. et al. BERMUDA: a novel deep transfer learning method for single-cell RNA sequencing batch correction reveals hidden high-resolution cellular subtypes. Genome Biol. 20, 1–15 (2019).
Menden, K. et al. Deep learning–based cell composition analysis from tissue expression profiles. Sci. Adv. 6, aba2619 (2020).
Parmar, N. et al. Image transformer. In Proc. 35th International Conference on Machine Learning Vol. 80, 4055–4064 (PMLR, 2018); https://proceedings.mlr.press/v80/parmar18a.html
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Vol. 1, 4171–4186 (Association for Computational Linguistics, 2018).
Le, Q. V. et al. XLNet: generalized autoregressive pretraining for language understanding. In Advances in Neural Information Processing Systems Vol. 32 (NeurIPS 2019); https://proceedings.neurips.cc/paper/2019/hash/dc6a7e655d7e5840e66733e9ee67cc69-Abstract.html
Yin, P., Neubig, G., Yih, W. & Riedel, S. TaBERT: pretraining for joint understanding of textual and tabular data. In Proc. 58th Annual Meeting of the Association for Computational Linguistics 8413–8426 (Association for Computational Linguistics, 2020); https://doi.org/10.18653/V1/2020.ACL-MAIN.745
Du, J. et al. Gene2vec: distributed representation of genes based on co-expression. BMC Genomics 20, 7–15 (2019).
Cortal, A., Martignetti, L., Six, E. & Rausell, A. Gene signature extraction and cell identity recognition at the single-cell level with Cell-ID. Nat. Biotechnol. 39, 1095–1102 (2021).
Kiselev, V. Y., Yiu, A. & Hemberg, M. scmap: Projection of single-cell RNA-seq data across data sets. Nat. Methods 15, 359–362 (2018).
Guo, H. & Li, J. scSorter: assigning cells to known cell types according to marker genes. Genome Biol. 22, 1–18 (2021).
Choromanski, K. et al. Rethinking attention with performers. In International Conference on Learning Representations (NIPS, 2021).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 1–19 (2019).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 1–12 (2017).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360.e4 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabol. 24, 593–607 (2016).
Xin, Y. et al. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metabol. 24, 608–615 (2016).
Nica, A. C. et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 23, 1554–1562 (2013).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Nat. Precedings https://doi.org/10.1038/npre.2010.4282.1 (2010).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Hwang, S. et al. HumanNet v2: human gene networks for disease research. Nucl. Acids Res. 47, D573–D580 (2019).
Liu, T.-Y. et al. Do transformers really perform badly for graph representation? In Advances in Neural Information Processing Systems Vol. 34 (NeurIPS, 2021).
Yun, S., Jeong, M., Kim, R., Kang, J. & Kim, H. J. Graph transformer networks. In 33rd Conference on Neural Information Processing Systems (NeurIPS, 2019).
McDavid, A. et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013).
Goldberg, Y. Neural Network Methods for Natural Language Processing Vol. 10, 1–311 (Springer, 2017); https://doi.org/10.2200/S00762ED1V01Y201703HLT037
Zhang, Y., Jin, R. & Zhou, Z.-H. Understanding bag-of-words model: a statistical framework. Int. J. Mach. Learn. Cybernetics 1, 43–52 (2010).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742 (2014).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 46 (2019).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 1–21 (2018).
Litviňuková, M. et al. Cells of the adult human heart. Nature 588, 466–472 (2020).
Tucker, N. R. et al. Transcriptional and cellular diversity of the human heart. Circulation 142, 466–482 (2020).
Lukassen, S. et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 39, e105114 (2020).
He, S. et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 21, 1–34 (2020).
Zhang, X. et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucl. Acids Res. 47, D721–D728 (2019).
Kimmel, J. C. & Kelley, D. R. Semi-supervised adversarial neural networks for single-cell classification. Genome Res. 31, gr.268581.120 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 1–9 (2019).
Yang, F. et al. scBERT as a Large-scale Pretrained Deep Language Model for Cell Type Annotation of Single-cell RNA-seq (Zenodo, 2022); https://doi.org/10.5281/zenodo.6572672
Acknowledgements
We thank B. Jiang and Y. Ji for their valuable suggestions on model building and experimental design. We thank T. Shen for advice on the large-scale model pretraining. H.L. was supported by the National Key R&D Program of China (grant no. 2018YFC0910500), a SJTU-Yale Collaborative Research Seed Fund, and Neil Shen’s SJTU Medical Research and Key-Area Research. F.Y. was supported by Development Program of Guangdong Province (grant no. 2021B0101420005).
Author information
Authors and Affiliations
Contributions
F.Y. and J.Y. conceived and designed the project. W.W. developed and implemented the algorithms under the guidance of F.Y. and J.Y.. W.W. and F.W. collected the datasets. W.W., F.Y. and F.W. conducted the experiments, data analysis and method comparisons. F.Y. and W.W. drew the figures and wrote the manuscript, with the guidance of J.Y. and H.L. Y.F. and F.W. finalized the manuscript and figures. D.T. gave suggestions for the design of the Transformer architecture, and the application of the NLP technology. J.H. gave suggestions on improving the manuscript. F.Y. and F.W. revised the figures and manuscript. All of the authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Jesper Tegner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The system analysis of the architecture design of scBERT.
a, Performance of scBERT (with/without pre-training) measured by accuracy and F1-score on Zheng68K dataset using 5-fold cross-validation. scBERT with pre-training is trained on over 1,000,000 cells from public scRNA-seq data from PanglaoDB. In the contrast, the model weights of scBERT without pre-training are initiated randomly. Box plot shows the median (centre lines), interquartile range (hinges) and 1.5 times the interquartile range (whiskers). b, Performance evaluation on the effect of gradually removing marker genes (no deletion, deletion of 10%, deletion of 50% and deletion of 100% markers) on accuracy. Box plot shows the median (centre lines), interquartile range (hinges), and 1.5 times the interquartile range (whiskers). The green dashed line represents the best performance achieved by other cell type annotation methods with all marker genes. c, UMAP representation of alpha, beta, delta, and gamma cells from Muraro dataset coloured by gene2vec embedding (sum of 200-dimension vectors) (top) and scBERT embedding (bottom) of alpha-specific gene LOXL4. d, The heatmap of average attention matrix obtained by taking an element-wise average across all attention matrices in multi-head multi-layer Performers. Each value \(A\left( {i,j} \right)\) (i and j indicate the index of row and column) represents how much attention from gene i was paid to gene j. e, Sensitivity analysis of hyperparameters includes the number of bins (top left), the dimension of scBERT embedding vector (top right), the number of attention heads (bottom left) and the number of Performer encoder layers (bottom right).
Extended Data Fig. 2 Performance comparison between scBERT and other cell type annotation methods on intra-datasets.
a, Performance of scBERT and other automatic cell type annotation methods measured by F1-score on n = 6 datasets (Zheng68K, Baron, Muraro, Xin, Segerstolpe, and MacParland) using 5-fold cross-validation. Box plots show the median (centre lines), interquartile range (hinges), and 1.5 times the interquartile range (whiskers). b, Performance of scBERT and marker-based methods (SCINA, Garnett, scSorter) measured by accuracy (left) and F1-score (right) on Zheng68K dataset using 5-fold cross-validation. Box plot shows the median (centre lines), interquartile range (hinges), and 1.5 times the interquartile range (whiskers). c-d, Performance of scBERT and other automatic cell type annotation methods measured by accuracy (c) and F1-score (d) on n = 3 datasets (Tucker dataset, lung dataset and Human Cell Atlas dataset) using 5-fold cross-validation. Box plots show the median (centre lines), interquartile range (hinges), and 1.5 times the interquartile range (whiskers).
Extended Data Fig. 3 Heatmaps for the confusion matrices of the results on Zheng68k dataset for other comparison methods.
a, The tSNE plots show the cell type annotation results of comparison methods (scNym, SciBet, Seurat, SingleR, CellID_cell, CellID_group, scmap_cell, scmap_cluster, SCINA, Garnett, scSorter) on Zheng68K dataset. The colours indicate the cell types annotation result from each individual method.
Extended Data Fig. 4 t-SNE plots of the cell type annotation results on Zheng68K dataset (n = 68,450 cells).
a, Heatmaps for the prediction confusion matrices on Zheng68K dataset for scNym, SciBet, SingleR, CellID_group, scmap_cell, and scmap_cluster. b, Heatmaps for the prediction confusion matrices on the imbalanced dataset constructed from Zheng68K dataset for Seurat, SingleR, CellID_cell, CellID_group, scmap_cell, and scmap_cluster.
Extended Data Fig. 5 Performance comparison between scBERT and other cell type annotation methods on cross-cohort dataset and cross-organ dataset.
a, t-SNE representation of alpha, beta, delta, and gamma cells from four pancreas datasets (n = 10,220 cells). The top left t-SNE plot is coloured by the annotated cell types provided by the atlas from the original paper, meanwhile other t-SNE plots are coloured by the cell type annotation results of comparison methods (SciBet, Seurat, SingleR, CellID_cell, CellID_group, scmap_cell, and scmap_cluster). b, Performance of scBERT and other cell type annotation methods measured by accuracy (left) and F1-score (right) on datasets from 3 organs (n = 17,384) using 5-fold cross-validation. Box plots show the median (centre lines), interquartile range (hinges), and 1.5 times the interquartile range (whiskers).
Extended Data Fig. 6 The distribution of the top attention sum genes across the four cell types of the Muraro dataset.
a, UMAP representation of alpha, beta, delta, and gamma cells from Muraro dataset coloured by expression distribution of top attention sum genes that are consistent with reported marker genes for alpha, beta, delta and gamma cells, respectively. b, UMAP representation of alpha, beta, delta, and gamma cells from Muraro dataset coloured by expression distribution of top attention sum genes that have distinguishing patterns on corresponding cell types but have not been reported as markers yet.
Supplementary information
Supplementary Data 1
Supplementary Tables 1–18.
Source data
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, F., Wang, W., Wang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat Mach Intell 4, 852–866 (2022). https://doi.org/10.1038/s42256-022-00534-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00534-z
This article is cited by
-
AI learns from chromatin data to uncover gene interactions
Nature (2025)
-
DNA promoter task-oriented dictionary mining and prediction model based on natural language technology
Scientific Reports (2025)
-
scCompressSA: dual-channel self-attention based deep autoencoder model for single-cell clustering by compressing gene–gene interactions
BMC Genomics (2024)
-
Pathogenomics for accurate diagnosis, treatment, prognosis of oncology: a cutting edge overview
Journal of Translational Medicine (2024)
-
STASCAN deciphers fine-resolution cell distribution maps in spatial transcriptomics by deep learning
Genome Biology (2024)