Abstract
Accurate prediction of damaging missense variants is critically important for interpreting a genome sequence. Although many methods have been developed, their performance has been limited. Recent advances in machine learning and the availability of large-scale population genomic sequencing data provide new opportunities to considerably improve computational predictions. Here we describe the graphical missense variant pathogenicity predictor (gMVP), a new method based on graph attention neural networks. Its main component is a graph with nodes that capture predictive features of amino acids and edges weighted by co-evolution strength, enabling effective pooling of information from the local protein context and functionally correlated distal positions. Evaluation of deep mutational scan data shows that gMVP outperforms other published methods in identifying damaging variants in TP53, PTEN, BRCA1 and MSH2. Furthermore, it achieves the best separation of de novo missense variants in neuro developmental disorder cases from those in controls. Finally, the model supports transfer learning to optimize gain- and loss-of-function predictions in sodium and calcium channels. In summary, we demonstrate that gMVP can improve interpretation of missense variants in clinical testing and genetic studies.
Missense variants are major contributors to genetic risk of cancers1,2 and developmental disorders3–5. Missense variants have been used, along with protein-truncating variants, to implicate new risk genes and are responsible for many clinical genetic diagnoses; however, the majority of rare missense variants are probably benign or only have minimal functional impact. As a result of the uncertainty of the functional impact, most rare missense variants reported in clinical genetic testing are classified as variants of uncertain significance6, leading to ambiguity, confusion, overtreatment and missed opportunities for clinical intervention. In human genetic studies to identify new risk genes by rare variants, pre-selecting damaging missense variants on the basis of computational prediction is a necessary step to improve statistical power4,5,7,8. Computational methods are therefore critically important to interpret missense variants in clinical genetics and disease gene discovery studies.
Numerous methods such as Polyphen9, SIFT10, CADD11, REVEL12, MetaSVM13, M-CAP14, Eigen15, MVP16, PrimateAI17, model predictive control (MPC)18 and correct classification rates (CCR)19 have been developed to address the problem. These methods differ in several aspects such as the prediction features, how the features are represented in the model, the training datasets and how the model is trained. Sequence conservation or local protein structural properties are the main prediction features for early computational methods such as GERP20 and PolyPhen. The MPC and CCR methods estimate sub-genic coding constraints from large human population sequencing data, providing additional information not captured by past methods. PrimateAI learns the protein context from sequences and local structural properties using deep representation learning. A number of studies have reported evidence that functionally damaging missense variants are clustered in three-dimensional protein structures21–23. Co-evolution captures the functional correlation between positions. Recent studies24,25 have shown that co-evolution helps to improve the prediction accuracy.
Here we present the graphical missense variant pathogenicity predictor (gMVP), a graph attention neural network model designed to effectively represent or learn the representation of all of the information sources to improve prediction of the functional impact of missense variants. gMVP uses a graph to represent a variant and its protein context, with node features describing sequence conservation and local structural properties; it also uses a graph attention neural network to learn the representation of a large protein context and uses the co-evolution strength as edge features that can potentially pool information about conservation and coding constraints of distant but functionally correlated positions. We trained gMVP using curated pathogenic variants and random rare missense variants in the human population. We then benchmarked the performance using datasets that have been curated or collected by entirely different approaches, for example: cancer somatic mutation hotspots26; functional readout datasets from deep mutational scan studies of well-known risk genes27–30; and de novo missense variants (DNMs) from studies of autism spectrum disorder (ASD)4 and neurodevelopmental disorder (NDD)5. Finally, we investigated the potential utility of transfer learning for classifying gain-of-function (GOF) and loss-of-function (LOF) variants in specific gene families based on the generic model trained across all genes.
Results
Model architecture and prediction features
gMVP is a supervised machine learning method for predicting functionally damaging missense variants. The functional consequence of missense variants depends on both the type of amino acid substitution and its protein context. gMVP uses a graph attention neural network to learn representation of protein sequence and structure context and context-dependent impact of amino acid substitutions on protein function.
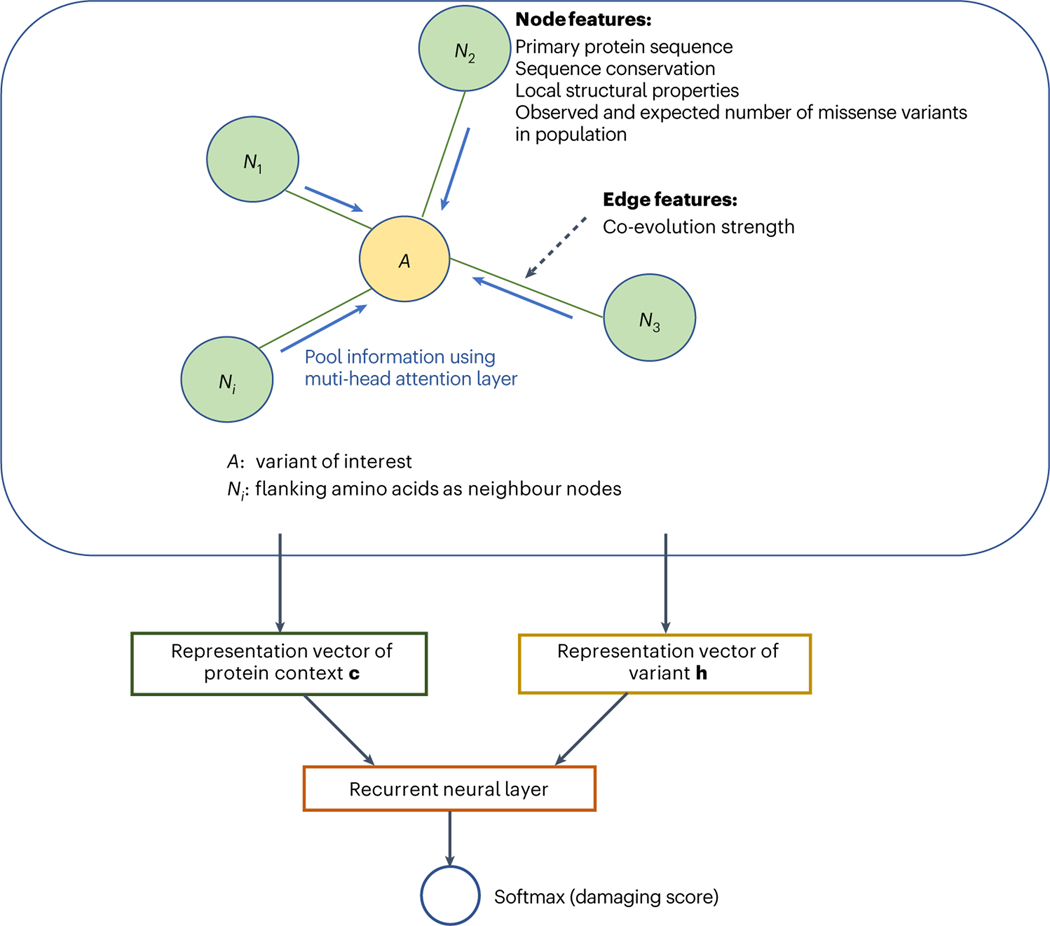
The main component of gMVP is a graph that represents a variant and its protein context (Fig. 1 and Supplementary Fig. 1). Given a variant, we define the 128 amino acids flanking the reference amino acid as protein context. We note that the average length of a protein domain annotated in the UniProt database is about 110 amino acids (Supplementary Fig. 8). We build a star-like graph with the reference amino acid as the centre node and the flanking amino acids as context nodes and connect the centre node and every context node with edges. We use co-evolution strength between the centre node of the variant and the context node as edge features. The co-evolution strength is highly correlated with functional interactions and protein residue–residue contact that captures the potential three-dimensional neighbours in folded proteins24,25,31,32. This architecture therefore allows the model to directly represent interactions between the position of interest and each flanking position in a wide context window. For the centre node, we include the amino acid substitution, evolutionary sequence conservation, and predicted local structural properties, such as secondary structures, as features (Methods). For context nodes, in addition to primary sequence, sequence conservation and local structure features, we also include the expected and observed number of rare missense variants in the human population to capture the selection effect of damaging variants in humans18,19. Let and denote input feature vectors for the centre node, neighbour nodes and edges, respectively. We first use three one-depth dense layers to encode and to latent representation vectors and , respectively. We then use a multi-head attention layer to learn the attention weight for each neighbour and to learn a context vector by weighting the neighbours. Attention scores play a key part in attention-based neural networks33,34. Our attention scores account for both the node features and the edge features. Specifically, we use tanh as attention scores, where tanh denotes a hyperbolic tangent activation function, and is the weight matrix to be trained. We next used a gated recurrent layer35—which is widely used to leverage sequence context in natural language modelling—to integrate vectors and of the variant. Finally, we use a linear layer and a sigmoid layer to perform classification and output the damaging scores.
Fig. 1 |. An overview of gMVP model.
gMVP uses a graph to represent a variant and its protein context defined as 128 amino acids flanking the reference amino acid. The amino acid of interest is the centre node (coloured orange) and the flanking amino acids are the context nodes (coloured light green). All context nodes are connected with the centre node but not each other. The edge feature is co-evolution strength. The node features include conservation and predicted structural properties. Centre node features also include the amino acid substitution; context node features include the primary sequence and the expected and observed number of rare missense variants in human population. We use three one-depth dense layers to encode the input features to latent representation vectors and used a multi-head attention layer to learn context vector c. We then use a recurrent neural layer connected with softmax layer to generate prediction score from c and the representation vector h of variant.
Model training and testing
We collected likely pathogenic and benign missense variants from curated databases (HGMD36, ClinVar37 and UniProt38) as training positives and negatives, respectively, and excluded the variants with conflicting evidence in the databases (Methods). To balance the positive and negative sets, we randomly selected rare missense variants observed in human population sequencing data DiscovEHR as additional negatives for training. In total there are 59,701 positives and 59,701 negatives, which cover 3,463 and 14,222 genes, respectively. We used a stochastic gradient descent algorithm39 to update the model’s parameters at an initial learning rate of 1 × 10−3 and applied early stopping with validation loss as a metric to avoid overfitting. We implemented the model and training algorithms using TensorFlow40. The whole training process took ~4 h on a Linux workstation with one NVIDIA Titan RTX GPU. When benchmarking the performance using a range of datasets, we compared gMVP with other widely used methods in genetic studies such as PrimateAI17, M-CAP14, CADD11, MPC18, REVEL12, MVP16, ClinPred41 and BayesDel42.
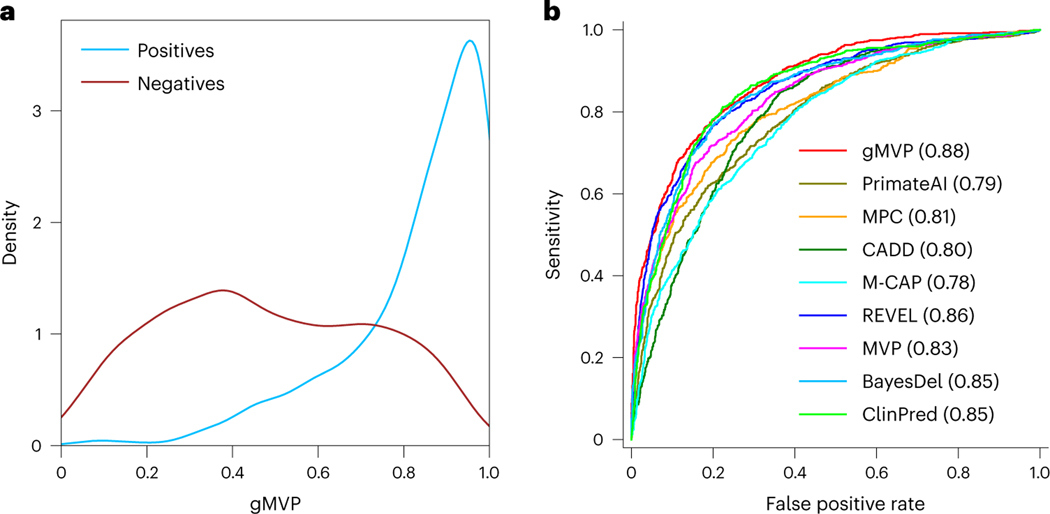
Human-curated pathogenic variants have hidden false positives that are probably caused by systematic biases and errors, which can be picked up by deep neural networks; therefore, conventional approaches for performance evaluation, using testing data randomly partitioned from the same source as the training data, usually lead to an inflated performance measure. To objectively evaluate the performance of the model, we compiled cancer somatic mutations that are unlikely to share the same systematic errors as the training datasets. We included missense mutations located in inferred hotspots on the basis of statistical evidence from a recent study26 as positives and randomly selected rare variants from the DiscovEHR database43 as negatives. The gMVP score distributions of cancer hotspot mutations and random variants have distinct modes (Fig. 2a). We selected a threshold of 0.75 to indicate a binary prediction for other downstream analyses that can best separate the score distributions of the positives and negatives. When compared with other published methods, gMVP achieved the best performance with an area under the receiver operating characteristic curve (AUROC) of 0.88 (Fig. 2b and Supplementary Table 2). REVEL is close with an AUROC of 0.86.
Fig. 2 |. Evaluating gMVP and published methods using cancer somatic mutation hotspots and random variants in population.
a, The gMVP score distributions for variants in cancer hotspots (labelled positives) and random missense variants in population (labelled negatives). b, Comparisons between the ROC curves of gMVP and other published methods. The ROC curves are evaluated on 878 cancer mutations located in hotspots from 209 genes and 1,756 (that is, a twofold greater number of positives) randomly selected rare variants from the DiscovEHR data.
gMVP can identify damaging variants in known disease genes
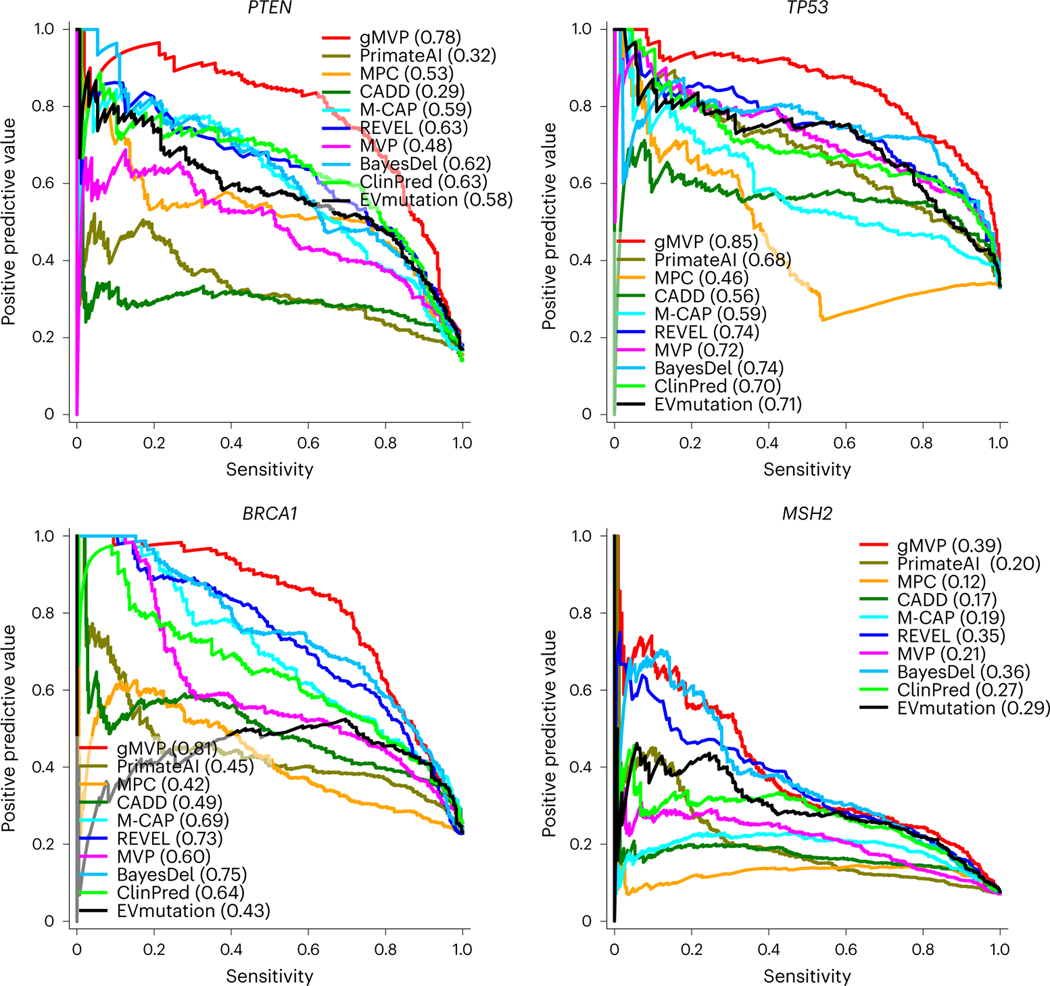
Missense variants that occur in different protein contexts—even in the same gene—can have different impacts. This is the core problem in interpreting variants from known risk genes in clinical genetic testing and the discovery of new disease genes. As performance evaluation using variants across genes is confounded by gene-level properties, here we aim to evaluate the ability of gMVP and other methods to distinguish damaging variants from neutral variants in the same genes. To this end, we obtained functional readout data from deep mutational scan assays of four well-known disease risk genes, TP5330, PTEN29, BRCA128 and MSH227, as benchmark data. The data include 432 damaging (positives) and 1,476 neutral (negatives) variants for BRCA1; 262 positives and 1,632 negatives for PTEN; 540 positives and 1,108 negatives for TP53; and 414 positives and 5,439 negatives for MSH2, respectively. We note that all variants in these four genes were excluded during gMVP training to avoid inflation in performance evaluation.
We first investigated the gMVP score distributions of damaging and neutral variants. Damaging variants clearly have different score distribution compared with the neutral variants in each gene (Supplementary Fig. 2). gMVP scores are also highly correlated with functional scores from the deep mutational scan assays, with a Spearman correlation coefficient of 0.67 (P = 1 × 10–246), −0.48 (P = 8 × 10–122), −0.53 (P = 7 × 10–51) and 0.29 (P = 7 × 10–117) in TP53, PTEN, BRCA1 and MSH2, respectively (Supplementary Fig. 3 and Supplementary Table 3–6).
We then used functional readout data as the ground truth to estimate precision–recall and compared gMVP with other methods. The areas under the precision–recall curves (AUPRCs) of gMVP are 0.78, 0.85, 0.81 and 0.39 for PTEN, TP53, BRCA1 and MSH2, respectively (Fig. 3), whereas the AUPRCs of the second-best method (REVEL) are 0.63, 0.74, 0.73 and 0.35, respectively. PrimateAI, a recent deep representation learning-based method, has AUPRCs of 0.32, 0.68, 0.45 and 0.20, respectively. A comparison using receiver operating characteristic (ROC) curves shows similar patterns (Supplementary Fig. 4).
Fig. 3 |. Evaluating gMVP and published methods in identifying damaging variants in known disease genes such as TP53, PTEN, BRCA1 and MSH2.
The precision–recall curves of gMVP and published methods are shown for each gene using functional readout data—labelled on the basis of the recommended threshold—as the ground truth.
Prioritizing rare DNMs using gMVP
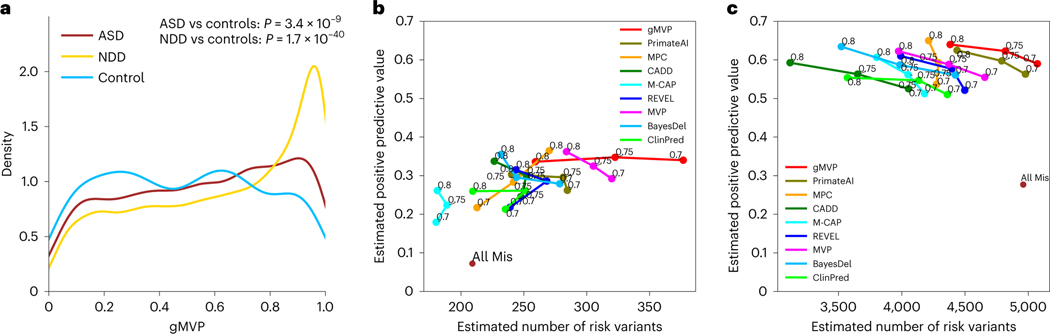
To further evaluate the utility of gMVP in new risk gene discovery, we compared the gMVP scores of DNMs from cases with developmental disorders with those from controls. We obtained published DNMs from 5,924 cases in an ASD study4, from 31,058 cases in an NDD study5 and from 2,007 controls (unaffected siblings from the ASD study)4. Although there is no ground truth because most of these DNMs were not previously implicated with diseases, there is a substantial excess of such variants in cases compared with the controls3,44,45, suggesting that a substantial fraction of variants in cases are pathogenic. We therefore tested whether the predicted scores of variants in cases and controls are significantly different and used significance as a proxy of performance (Fig. 4a). gMVP achieves a P-value of 38 × 10–9 and 28 × 10–40 for ASD or NDD versus controls, respectively, whereas the second-best method PrimateAI achieves a P-value of 38 × 10–6 and 28 × 10–38, respectively (Supplementary Fig. 5).
Fig. 4 |. Evaluating gMVP and published methods in distinguishing rare DNMs in cases with neurodevelopmental disorders from those in controls.
a, Distributions of gMVP-predicted scores for rare DNMs from ASD and NDD cases, as well as controls. We used a two-sided Mann–Whitney U test to assess the statistical significance of the difference between the cases and controls. Controls are unaffected siblings from the ASD study. b, Comparisons between gMVP and other published methods using DNMs from ASD cases and controls by precision–recall–proxy curves. Numbers on each point indicate rank percentile thresholds. The positions of the All Mis points are estimated from all missense variants without using any prediction method. c, The same comparison using data from NDD cases and controls.
We then calculated the enrichment rate of predicted damaging DNMs of a method with a certain threshold in cases compared with the controls, and then estimated the precision and the number of true risk variants (Methods), which is a proxy of recall because the total number of true positives in all cases is a (unknown) constant that is independent of the methods. The estimated precision and recall values are directly related to the power of detecting new risk genes5,46. We also calculated the estimated precision and number of true risk variants on all missense variants (denoted as All Mis) in the dataset, without using any predictor. We compared the performance of gMVP with other methods by the estimated precision and recall–proxy curves (Fig. 4b,c). The optimal threshold of the gMVP rank score in cancer hotspots is 0.75; with this, we observed an enrichment rate of 2.7 and 1.5 in NDD and ASD, respectively (Supplementary Tables 7 and 8), which corresponds to an estimated precision–recall of (0.62, 4,818) and (0.35, 328), respectively. Furthermore, when using a lower threshold of 0.7, gMVP can still keep the precision as high as 0.34, and achieved a recall of 377 in ASD. PrimateAI achieves overall second-best estimated precision and recall under different thresholds in both ASD and NDD. MPC, with a threshold of 0.8, can reach a high precision at 0.65 and 0.36 in NDD and ASD, respectively, but overall it has substantially lower recall than gMVP and PrimateAI.
Classifying mode of action of variants via transfer learning
In many genes, the functional impact of missense variants is complex and cannot be simply captured by a binary prediction. Heyne et al.47 recently investigated the pathogenetic variants that alter the channel activity of voltage-gated sodium and calcium channels and inferred LOF and GOF variants on the basis of clinical phenotypes of variant carriers and electrophysiology data. The study also described a computational model (funNCion) to predict LOF and GOF variants using a large number of human-curated features on electrophysiological properties. Here we sought to classify LOF and GOF variants using the gMVP model through transfer learning without additional curated prediction features. Transfer learning allows us to further train a model for a specific purpose using a limited number of training points by only exploring a reasonable subspace of the whole parameter spaces guided by previously trained models.
We obtained 1,517 pathogenetic and 2,328 neutral variants in ten voltage-gated sodium and ten calcium channel genes, in which 518 and 309 variants were inferred as LOF and GOF variants, respectively, from the work by Heyne and colleagues47. To benchmark the performance, we used the same training and testing sets (90/10% breakdown) as funNCion.
We first evaluated the performance of gMVP and previous methods in distinguishing LOF or GOF from neutral variants. gMVP and REVEL both achieved the best AUROC of 0.94 (Fig. 5a and Supplementary Table 9). FunNCion47, which was trained specifically on the variants of the ion-channel genes, achieved a nearly identical AUROC of 0.93. We next sought to improve the performance using transfer learning. Starting with the weights from the original gMVP model, we trained a new model, gMVP-TL1, with both LOF and GOF variants in these genes as positives, and neutral variants as negatives (Methods). gMVP-TL1 achieved an AUROC of 0.96, outperforming the original gMVP and published methods. Furthermore, to distinguish LOF and GOF variants, we trained another model, gMVP-TL2, also starting with the weights of the original gMVP model, but with different output labels for training (LOF versus GOF; Methods). The training set includes 465 LOF and 279 GOF variants, whereas the testing set comprises 51 LOF and 30 GOF variants. gMVP-TL2 achieved an AUROC of 0.95, substantially better than funNCion (AUROC, 0.84), which trained on the same variants set with manually curated prediction features (Fig. 5b and Supplementary Table 10). This demonstrates that the gMVP model aided by transfer learning technique can accurately predict GOF and LOF variants in channel genes with a very limited training dataset.
Fig. 5 |. Evaluating gMVP and other published methods in classifying pathogenetic and neutral variants, and in predicting GOF and LOF variants in ion-channel genes.
a, Comparison of ROC curves in classifying pathogenic variants and neutral variants. gMVP-TL1 denotes the model further trained on the pathogenetic and neutral variants in SCNxA genes starting from the weights of the original gMVP model. b, Comparison of ROC curves in classifying GOF and LOF variants. gMVP-TL2 denotes the model further trained on GOF and LOF variants starting from the weights of the original gMVP model.
gMVP captures conservation, structure and selection in humans
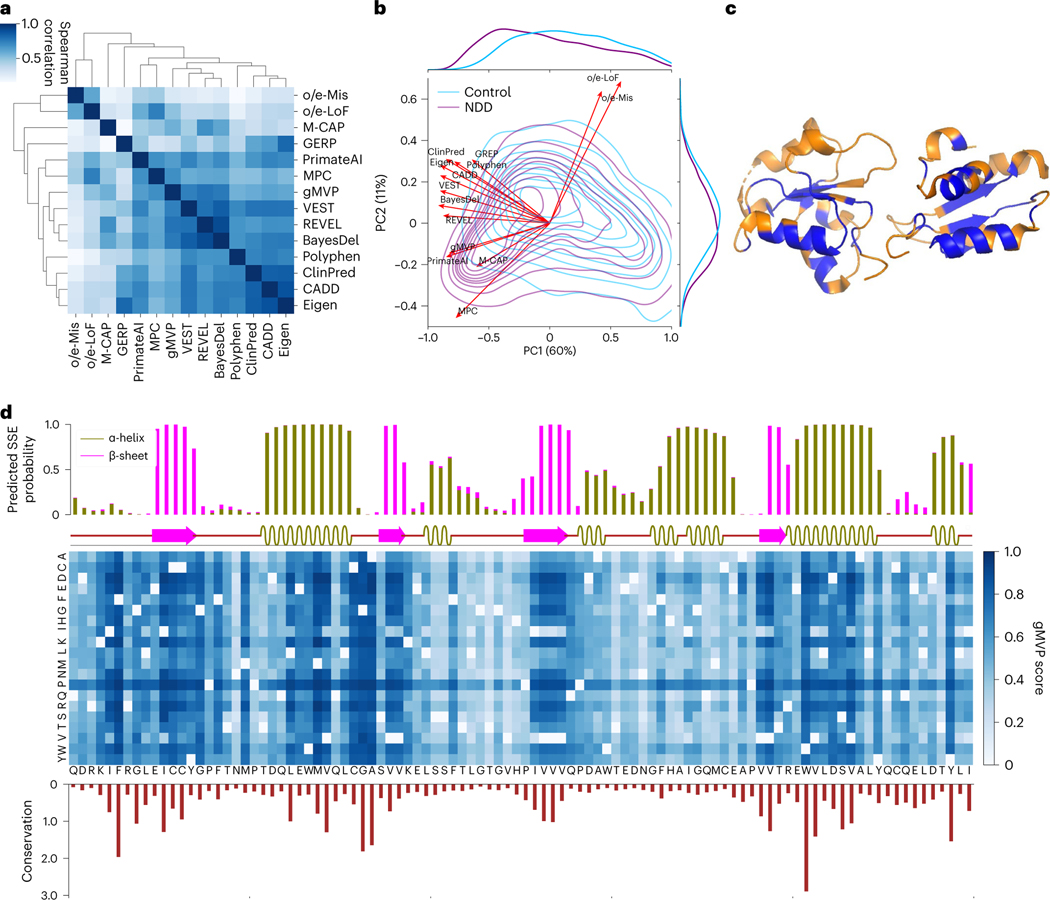
We calculated the correlation between predicted scores of gMVP and other methods on DNMs from ASD and NDD cases and controls (Fig. 6a). gMVP has the highest correlation with REVEL (Spearman ρ = 0.78), followed by a few other widely used methods such as BayesDel, MPC, CADD and PrimateAI (ρ > 0.6).
Fig. 6 |. Interpreting gMVP predictions with conservation, protein structure and genetic coding constraints.
a, Spearman correlation between gMVP and other published methods, calculated by scores of the DNMs in ASD, NDD and controls. b, PCA on DNMs from ASD and NDD cases and controls. Red arrows show the loadings of gMVP and published methods on the first two components; the density contour shows the distribution of PC1/2 scores of the variants in NDD and controls. The density curves along the axes show the distribution of PC1 or PC2 scores of the cases and controls. c, The protein tertiary structure of BRCT2 domain of BRCA1. We coloured a residue blue if at least one missense on this position is predicted to be damaging (gMVP > 0.75) and orange otherwise. d, gMVP scores of all possible missense variants on the BRCT2 domain of BRCA1. The top bar plot shows the predicted probabilities of the protein secondary structures, whereas the bar below shows the real protein secondary structures calculated by DSSP. The middle heat map shows gMVP scores for all possible missense variants on each protein position (the darker the colour, the higher the gMVP score). The bottom histogram shows the evolutionary conservation measured with the Kullback–Leibler divergence between amino acid distribution among homologous sequences and amino acid distribution in nature.
We then performed principal component analysis (PCA) on the DNMs from cases and controls to investigate the contributing factors that separate the variants (Fig. 6b and Supplementary Fig. 6). The input of the PCA is a score matrix in which rows represent variants and columns represent predicted scores by gMVP and other methods. We included two additional columns with gene-level gnomAD constraint metrics o/e-LoF and o/e-Mis (observed number over expected number for LOF and missense)48 to represent selection effect in humans. The first component (PC1) captures the majority of the variance of the data and best separates the DNMs in cases and the ones in controls. All methods have large loadings on PC1 (Fig. 6b). The second component (PC2) is largely driven by the gene-level gnomAD constraint metrics (Fig. 6b). The joint distribution of PC1/2 scores of DNMs from controls has a single mode at the centre. The joint distributions of scores of DNMs from cases have two modes (Fig. 6b and Supplementary Fig. 6b) that represent mixtures of likely pathogenic variants and random DNMs. Notably, gnomAD metrics have near orthogonal loadings on PC1/2 with GERP, which is purely based on cross-species conservation, suggesting that selection effect in humans provides complementary information to evolutionary conservation about genetic effect of missense variants. All methods (PolyPhen, eigen, CADD, VEST and REVEL) that do not use human or primate population genome data have loadings close to GERP on PC1/2. MPC and M-CAP, which use sub-genic or gene-level mutation intolerance metrics similar to gnomAD metrics, have the closest loadings as gnomAD metrics on PC1/2. gMVP and PrimateAI have similar loadings that are in the middle of GERP and gnomAD metrics.
We inspected the BRCT2 domain of BRCA1 to show how the gMVP model captures context-dependent functional impact. We observed that most damaging variants predicted by gMVP (>0.75) are located in the core region of BRCT2 domain (Fig. 6c). Furthermore, gMVP scores are highly correlated with evolutionary conservation (Fig. 6d and Supplementary Fig. 7a; ρ = 0.57). Variants in the β-sheets are much more damaging than the ones in α-helix regions, and the ones in α-helix regions are more damaging than the ones in coil regions (Fig. 6d and Supplementary Fig. 7b), consistent with past discoveries21,49,50. Finally, amino acids mutated to proline (P) in helix regions are predicted to be highly damaging, even in positions not well conserved (Fig. 6d). This is consistent with the fact that proline rarely occurs in the middle of an α-helix51.
Discussion
We developed gMVP—a new method based on graph attention neural networks—to predict functionally damaging missense variants. gMVP uses attention neural networks to learn representations of protein sequence and structure context through supervised learning trained with large number of curated pathogenic variants. The graph structure allows co-evolution-guided pooling of predictive information of distal amino acid positions that are functionally correlated or potentially close in three-dimensional space. We demonstrated the utility of the gMVP in clinical genetic testing and new risk gene discovery studies. Specifically, we showed that gMVP achieves better accuracy in identification of damaging variants in known risk genes based on functional readout data from deep mutational scan studies. Furthermore, gMVP achieved better performance in prioritizing DNMs in cases with autism or NDD, suggesting that it can be used to pre-select damaging variants or weight variants to improve statistical power of new risk gene discovery. Finally, we showed that with transfer learning technique, gMVP model can accurately classify GOF and LOF variants in ion channels even with a limited training set without additional prediction features.
gMVP learns a representation of protein context from training data, whereas previous ensemble methods such as REVEL, M-CAP, MetaSVM and CADD used scores from other predictors or other human-engineered features as inputs. With recent progress of machine learning in protein structure prediction52–55, neural network representations could capture latent structure beyond common linear representations of understanding of the biophysical and biochemical properties. We showed that representation learning allows gMVP to capture the context-dependent impact of amino acid substitutions on protein function. PrimateAI is a recently published method that also uses deep representation learning. gMVP achieved better performance than PrimateAI in identification of damaging variants in known disease risk genes in comparisons that use functional readout data as well as in prioritizing rare DNMs from ASD and NDD studies. Although both models used evolutionary conservation and protein structural properties as features, the two methods have entirely different model architecture and training data. gMVP uses a graph attention neural network to pool information from both distal and local positions with co-evolution strength, whereas PrimateAI uses a convolutional neural network to extract local patterns from a protein context. For training data, gMVP used expert-curated variants and random variants in population as training positives and negatives, respectively. By contrast, PrimateAI used common variants in primates as negatives and unobserved variants in the population as positives. Based on functional readout data of the four well-known risk genes, only 15–25% of random variants have discernible impact on protein function. The positives used in PrimateAI training may therefore contain a large fraction of false positives. PrimateAI’s training strategy does have advantages. It avoids human interpretation bias and errors in curated databases of pathogenic variants, the positives used in gMVP training. It also can cover almost all human protein-coding genes, whereas curated databases such as ClinVar only cover hundreds of genes. Additionally, common variants in primates are probably all true negatives, whereas random observed rare variants in human population could have a non-negligible fraction of damaging variants. Making a new model that can use all of these datasets in training could further improve the prediction performance.
Several past studies have shown that the functional impact of missense variants is correlated among three-dimensional neighbours21,22,56. Pooling information from three-dimensional neighbours could therefore improve predictions of functional impact. However, directly considering three-dimensional distances is limited by the fact that most human proteins have no solved tertiary structures with considerable coverage. gMVP addresses this issue by taking a large segment of the protein context that include both local and distant positions that are potential neighbours in folded proteins, and then uses co-evolution strength to effectively pool information from potential three-dimensional neighbours. Used as edge features in a graph attention model, co-evolution strength allows more precise pooling of information from distant residues than the convolutional layer without prior structure. Co-evolution information has been used by previous methods for predicting functional impact of missense variants, such as PIVOTAL25, a supervised ensemble predictor that combines scores from existing methods and EVmutation, an unsupervised method that learns co-evolution and conservation using Markov random fields from multi-sequence alignments (MSAs). Moreover, co-evolution information has been used in ab initio protein structure prediction extensively32,54,57. The extraordinary performance of AlphaFold55,58 in CASP14 shows that it contains critical information about physical residue–residue distances for accurate structure prediction of most proteins in the human proteome. The language model Transformer33 has more recently been applied on protein sequences and MSAs to improve the performance of co-evolution strength estimation and protein residue–residue contacts prediction59–61. gMVP could be further improved by integrating components of Transformer and protein three-dimensional structures in the model. On the other hand, MSA-based methods are limited for the proteins with no or few homologous sequences and could be improved by integrating the learned representations on large-scale unlabelled sequence data using sequence language modelling60.
With transfer learning, the trained gMVP model can be further optimized for more specific tasks in genetic studies. The idea is to transfer the general knowledge learned from large training datasets to a new related and more specific task with only limited training data. The trained model can set the initial values of the weights in the model to be updated by further training to explore only a subspace of the whole parameter space. We have shown its feasibility in classifying GOF and LOF variants in the ion-channel genes using a limited number of training data points without additional prediction features. We expect that with transfer learning, gMVP can potentially improve variant interpretation by training gene family-specific models62 and identifying disease-specific damaging variants63.
Functional readout data from deep mutational scan provides strong evidence of classifying variants as damaging or neutral27–30,64,65. However, these in vitro functional readout assays usually reveal only one aspect of a protein’s function in a limited number of cell types; therefore, they are often not completely correlated with the functional impact of the variants in vivo. We expect that more comprehensive deep mutational scan assays will become available and facilitate substantial improvement in the training and evaluation of computational methods.
Finally, we showed that although evolutionary conservation remains one of the most informative sources for computational methods, selection in humans can provide complementary information for prediction. The selection coefficient is correlated with allele frequency, especially for variants under strong negative selection46,66–68. Larger population genome datasets can further improve estimation of allele frequency of rare variants. We anticipate large69 and diverse70 population genome data released in the future will improve estimation of selection effect in human and in turn improve gMVP.
Methods
Training datasets
For the positive training set, we collected: 22,607 variants from Clin-Var database37 under the pathogenic and likely pathogenic categories with a review status of at least one star; 48,125 variants from the Human Gene Mutation Database Pro v.2013 (HGMD) database36 under the disease mutation category; and 20,481 variants from UniProt labelled as disease-causing. For the negative training set, we collected 41,185 variants from ClinVar under the benign and likely benign categories, and 33,387 variants from SwissVar38 labelled as polymorphism. After excluding 3,751 variants with conflicting interpretations from the three databases, we have 63,304 and 66,102 unique positives and negatives, respectively. We next excluded 36,499 common variants (653 positives and 35,846 negatives) with an allele frequency >1 × 10–3 in gnomAD (all populations)48 and 3,080 overlapping variants (2,680 positives and 400 negatives) with testing datasets from the training dataset, resulting in a dataset of 59,701 positives and 29,856 negatives. To balance the positive and negative training samples, we randomly selected 29,845 rare missense variants from the DiscovEHR database43 that are not already covered by previously selected training data as additional negative training points. In the end we have 59,701 and 59,701 unique positive and negative training variants (Supplementary Table 1), which cover 3,463 and 14,222 genes, respectively.
Testing datasets
Cancer somatic mutation hotspots: we obtained 878 missense variants located in somatic missense mutations hotspots in 209 cancer driver genes from a recent study26 as positives, and randomly selected twofold more rare missense variants (N = 1,756) from the population sequencing data DiscovEHR43.
Functional readout data from deep mutational scan experiments: we compiled variants in BRCA128, PTEN29, TP5330 and MSH227. Findly and colleagues30 applied genome editing to measure the functional consequences of all possible single nucleotide variants (SNVs) in key regions of BRCA1, where the functional scores measured the SNV effects on the cell survival of the cloned cells. Mighell et al.29 used a yeast model to systematically evaluate the effect of PTEN mutations on lipid phosphatase activity in vivo. Kotler et al.30 created a synthetically designed library and measured the functional impact of the DNA-binding domain p53 variants in human cells in culture and in vivo. Jia et al.27 developed a human cell line model for MSH2 to measure the chemical selection for mismatch repair dysfunction. The functional scores for PTEN and BRCA1 correlate negatively, whereas the functional scores for TP53 and MSH2 correlate positively, with the pathogenicity of the variants, respectively. We used the suggested thresholds of the functional scores to label the positives and negatives for the variants. We only include the SNVs for comparison as most published methods do not provide scores for the non-SNVs. There are 432 positives and 1,476 negatives in BRCA1; 258 positives and 1,601 negatives in PTEN; 540 positives and 1,108 negatives in TP53; and 414 positives and 5,439 negatives in MSH2.
DNMs: to evaluate utility in new risk gene discovery, we used published rare germline DNMs from 5,924 cases and 2,007 controls in an ASD study4 and 31,058 cases in a neural developmental study5.
To fairly compare our methods with published methods, we excluded the overlapping variants with testing datasets from the training datasets. We further excluded all variants in PTEN, TP53, BRCA1 and MSH2 in training to avoid inflation in performance evaluation.
Past published methods included for comparison
We compared gMVP with PrimateAI, MPC, REVEL, M-CAP, MVP, ClinPred, BayesDel, EVmutation, SIFT, PolyPhen2, SIFT, phastCons71 and GERP. We calculated scores of EVmutation using its public software package (https://github.com/debbiemarkslab/EVmutation). We used the pre-computed scores of other methods compiled by dbNSFP. We annotated the variants in the testing test with these scores using VEP plug-in for dbNSFP.
The graph attention neural network model
gMVP uses a graph to represent a variant and its protein context. We first defined the 128 amino acids flanking the reference amino acid as protein context. We next built a star-like graph with the reference amino acid as the centre node and the flanking amino acids as context nodes, and with edges between the centre node and each context node (Fig. 1 and Supplementary Fig. 1).
Let , and denote input feature vectors for the centre node, each context node and each edge, respectively. We first used three one-depth dense layers to encode , and to latent representation vectors , and , respectively. We used RELU72 as the activation function and 512 neurons for each dense layer.
We then used a multi-head layer adapted from the attention layer in the Transformer model33 to pool information from context nodes and finally to learn a context vector . Specifically, for the kth head, we first calculated the value vectors for each context node by . We next calculated attention scores for each context node through , where tanh denotes a hyperbolic tangent activation function and is a position bias, which is a simplified positional encoding73. We note here that allows the model to capture local protein sequence context. Attention weights are calculated by applying a softmax operation on the attention scores,.
The context vector for the kth head is calculated as . The final context vector is obtained by a linear projection on the concatenation vector of the context vectors from each head,
Here denotes the number of heads and we used four heads in our model. And we note that in the model, , and are weight matrices to be trained.
We next used a gated recurrent unit layer35 to leverage the context vector and the latent vector of the given variant where the relative importance of the whole context can be determined. We used 512 neurons and a hyperbolic tangent activation function for the gated recurrent unit layer. We finally used a linear projection layer and a sigmoid layer to perform classification.
Input features
The centre node, which represents the variant, has the following features: reference and alternate amino acids, evolutionary conservation and predicted local structural properties. The context nodes have the following features: reference amino acids, evolutionary conservation, predicted local structural properties and observed and expected missense alleles in gnomAD48. The feature of edges is co-evolution strength between the position of variant and other positions, estimated from multiple sequence alignments of homologous sequences.
Reference and alternate amino acids (40 values): we used one-hot encoding with a dimension of 20 to represent reference and alternate amino acids.
Protein primary sequence (20 values): we also used one-hot encoding to represent each amino acid in the protein primary sequence.
Evolutionary conservation (42 values): we estimated the evolutionary conservation from two sources: (1) we searched the homologous of the protein of interest against SwissProt database74 with three iterations of search and then built the MSAs with HHblits suite75; (2) we downloaded the MSAs of 200 species from Ensembl website for each human protein sequence76. We then calculated the frequencies of 20 amino acids and the gap for each position for the two MSAs separately and concatenated the two frequency vectors.
Predicted protein structural properties (five values): we predicted the protein secondary structures (three values), solvent accessibility (one value) and the probability of a residue participating in interactions with other proteins (one value) using NetsurfP77.
Observed number of missense alleles in gnomAD and expected number (two values): to capture selection effect in human, we obtained the observed number of rare missense variants in gnomAD48 and the expected number of rare missense variants estimated using a background mutation model48.
Co-evolution strength (442 values): we extract pairwise statistics from the MSA as co-evolution strength. It is estimated based on the covariance matrix constructed from the input MSA. First, we compute one- and two-site frequency counts and , where A and B denote amino acid identities (20 + gap); δ is the Kronecker delta; and are position indexes on the aligned protein sequence; is the sequence index of the MSA with a total of aligned sequences; and indicates the amino acid identity of position on sequence . We then calculate the sample covariance (21 × 21) matrix and flatten it into a vector with 441 elements. We also convert the covariance matrix to a single value by computing its Frobenius norm and then concatenate the norm and the flattened vector as the edge features.
We built these features only for canonical transcripts defined by Ensembl78 v.92. We annotated the variants using VEP79.
Training algorithm
We used cross-entropy loss as the training loss. We used the Adam algorithm39 to update the model parameters with an initial learning rate of 1 × 10–3 and decayed the learning rate with a polynomial decay schedule80. We randomly selected 10% of training samples as validation set and early stopping was applied with validation loss as a watching metric. We trained five models by repeating the above training process five times, and for testing, we averaged the outputs of the five models as prediction scores. The model and training algorithm were implemented using TensorFlow40.
Classifying GOF and LOF variants using transfer learning.
To investigate the potential for transfer learning, we further trained gMVP to classify GOF and LOF variants in ion-channel genes with additional training data but without new features. We collected 1,517 pathogenetic and 2,328 neutral variants in SCNxA genes, which encode voltage-gated sodium and calcium channel proteins, in which 518 and 309 variants are inferred as LOF and GOF variants, respectively, from a recent study47.
We first trained a model, gMVP-TL1, to classify pathogenetic and neutral variants in SCNxA genes. We used the same dataset as funNCion47, including 3,466 variants for training and 379 variants for testing. We randomly selected 10% variants from training set as validation set. We used the same model architecture with gMVP and initialized weights of the new model with the weights of original gMVP model. In the new model training, we used Adam to update the parameters at an initial learning rate of 1 × 10–3 and used the validation loss as stopping criteria. We trained five gMVP-TL1 models, starting from each of the five trained gMVP models, and for testing, we averaged the outputs of these models as prediction scores.
We next trained another model gMVP-TL2 to classify GOF versus LOF variants in SCNxA genes. We used 744 variants as training set and 81 variants as testing set, which are the same sets used by funNCion47. Like gMVP-TL1, gMVP-TL2 were also trained starting from the weights of gMVP model previously trained using all genes. We used the same hyperparameter settings with gMVP-TL1 in training.
Normalization of scores using rank percentile
For each method, we first sorted predicted scores of all possible rare missense variants across all protein-coding genes and then converted the scores into rank percentiles. The higher rank percentile indicates more damaging, for example, a rank score of 0.9 indicates the missense variant is more likely to be damaging than 90% of all possible missense variants.
Precision–recall–proxy curves
As there are no ground-truth data to benchmark our performance on DNMs, we estimate precision and recall at various thresholds based on the enrichment of predicted damaging variants in cases compared to controls.
Let be the rate of synonymous variants in cases and be the rate of synonymous variants in controls. Then the synonymous rate ratio is defined as
Denote the total number of variants in cases as , the number of variants in controls as , the number of variants predicted as pathogenic in cases as and the number of variants predicted as pathogenic in controls as . We assume that for there to be no batch effect, the rate of synonymous variants should be the same in the cases and controls. So, we estimate the enrichment of predicted pathogenic variants in cases compared to controls by:
The true number of pathogenic DNMs is then estimated by
And the estimated precision is
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Pre-computed gMVP scores for all possible missense variants in canonical transcripts on human hg38 can be downloaded from https://www.dropbox.com/s/nce1jhg3i7jw1hx/gMVP.2021–02-28.csv.gz?dl=0. The training data of the main model were downloaded from http://www.discovehrshare.com/downloads (DiscovEHR), http://www.hgmd.cf.ac.uk/ac/index.php (HGMD), https://www.uniprot.org/docs/humpvar (UniProt) and https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/ (ClinVar). Other datasets supporting the findings of this study are available in the paper and the Supplementary Information.
Code availability
The codes for the model design and training and testing procedure are available on GitHub (https://github.com/ShenLab/gMVP/) and Zenodo81.
Supplementary Material
Acknowledgements
This work was supported by NIH grants (nos. R01GM120609, R03HL147197, U01HG008680 and K99HG011490) and the Columbia University Precision Medicine Joint Pilot Grants Program. We thank Y. Zhao, G. Zhong, M. AlQuraishi and D. Knowles for helpful discussions.
Footnotes
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42256–022-00561-w.
Peer review information Nature Machine Intelligence thanks Xiaoming Liu, Wim Vranken, Amit R Majithia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
References
- 1.Boettcher S et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang KL et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin SC et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 49, 1593–1601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satterstrom FK et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplanis J et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 586, 757–762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehm HL, Berg JS & Plon SE ClinGen and ClinVar—enabling genomics in precision medicine. Hum. Mutat. 39, 1473–1475 (2018). [Google Scholar]
- 7.He X et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 9, e1003671 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HT et al. Integrated Bayesian analysis of rare exonic variants to identify risk genes for schizophrenia and neurodevelopmental disorders. Genome Med. 9, 114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei I, Jordan DM & Sunyaev SR Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 10.1002/0471142905.hg0720s76 (2013). [DOI] [PMC free article] [PubMed]
- 10.Carter H, Douville C, Stenson PD, Cooper DN & Karchin R Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genom. 14, S3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ioannidis NM et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadeesh KA et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 48, 1581–1586 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Ionita-Laza I, McCallum K, Xu B & Buxbaum JD A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 48, 214–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi H et al. MVP predicts the pathogenicity of missense variants by deep learning. Nat. Commun. 12, 510 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundaram L et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 50, 1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samocha KE et al. Regional missense constraint improves variant deleteriousness prediction. Preprint at bioRxiv 10.1101/148353 (2017). [DOI]
- 19.Havrilla JM, Pedersen BS, Layer RM & Quinlan AR A map of constrained coding regions in the human genome. Nat. Genet. 51, 88–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davydov EV et al. Identifying a high fraction of the human genome to be under selective constraint using GERP plus. PLoS Comput. Biol. 6, e1001025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iqbal S et al. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc. Natl Acad. Sci. USA 117, 28201–28211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks M, Bartha I, di Iulio J, Venter JC & Telenti A Functional characterization of 3D protein structures informed by human genetic diversity. Proc. Natl Acad. Sci. USA 116, 8960–8965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivley RM, Dou XY, Meiler J, Bush WS & Capra JA Comprehensive analysis of constraint on the spatial distribution of missense variants in human protein structures. Am. J. Hum. Genet. 102, 415–426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopf TA et al. Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang S, Mort M, Stenson PD, Cooper DN & Yu H PIVOTAL: prioritizing variants of uncertain significance with spatial genomic patterns in the 3D proteome. Preprint at bioRxiv 10.1101/2020.06.04.135103 (2021). [DOI]
- 26.Chang MT et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia X et al. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk. Am. J. Hum. Genet. 108, 163–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Findlay GM et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mighell TL, Evans-Dutson S & O’Roak BJ A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype–phenotype relationships. Am. J. Hum. Genet. 102, 943–955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotler E et al. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell 71, 178–190.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 31.de Juan D, Pazos F & Valencia A Emerging methods in protein co-evolution. Nat. Rev. Genet. 14, 249–261 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Morcos F et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl Acad. Sci. USA 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaswani A et al. Attention is all you need. In 31st Conference on Neural Information Processing Systems 5998–6008 (NeurIPS, 2017). [Google Scholar]
- 34.Veličković P et al. Graph attention networks. In 6th International Conference on Learning Representations (Univ. Cambridge, 2018). [Google Scholar]
- 35.Cho K et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. In Proc. 2014 Conference on Empirical Methods in Natural Language Processing (Association for Computational Linguistics, 2014). [Google Scholar]
- 36.Stenson PD et al. Human gene mutation database (HGMD (R)): 2003 update. Hum. Mutat. 21, 577–581 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Landrum MJ et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucl. Acids Res. 42, D980–D985 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mottaz A, David FP, Veuthey AL & Yip YL Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics 26, 851–852 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kingma DP & Ba J Adam: a method for stochastic optimization. In 2015 International Conference on Learning Representations (ICLR, 2015). [Google Scholar]
- 40.Abadi M et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. Preprint at https://arxiv.org/abs/1603.04467 (2016).
- 41.Alirezaie N, Kernohan KD, Hartley T, Majewski J & Hocking TD ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am. J. Hum. Genet. 103, 474–483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng BJ PERCH: a unified framework for disease gene prioritization. Hum. Mutat. 38, 243–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dewey FE et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR Study. Science 354, aaf6814 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuk O et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl Acad. Sci. USA 111, E455–E464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heyne HO et al. Predicting functional effects of missense variants in voltage-gated sodium and calcium channels. Sci. Transl. Med. 12, eaay6848 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abrusán G & Marsh JA Alpha helices are more robust to mutations than beta strands. PLoS Comput. Biol. 12, e1005242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao M, Zhou H & Skolnick J Insights into disease-associated mutations in the human proteome through protein structural analysis. Structure 23, 1362–1369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S-C, Goto NK, Williams KA & Deber CM Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl Acad. Sci. USA 93, 6676–6681 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Senior AW et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Yang JY et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl Acad. Sci. USA 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Sun S, Li Z, Zhang R & Xu J Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Clarke D & Gerstein MB Leveraging protein dynamics to identify cancer mutational hotspots using 3D structures. Proc. Natl Acad. Sci. USA 116, 18962–18970 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anishchenko I, Ovchinnikov S, Kamisetty H & Baker D Origins of coevolution between residues distant in protein 3D structures. Proc. Natl Acad. Sci. USA 114, 9122–9127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tunyasuvunakool K et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao R et al. MSA transformer. In Proc. 38th International Conference on Machine Learning 8844–8856 (PMLR, 2021). [Google Scholar]
- 60.Rives A et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao R, Meier J, Sercu T, Ovchinnikov S & Rives A Transformer protein language models are unsupervised structure learners. In 2015 International Conference on Learning Representations (ICLR, 2015). [Google Scholar]
- 62.Lal D et al. Gene family information facilitates variant interpretation and identification of disease-associated genes in neurodevelopmental disorders. Genome Med. 12, 28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X et al. Disease-specific variant pathogenicity prediction significantly improves variant interpretation in inherited cardiac conditions. Genet. Med. 23, 69–79 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Starita LM et al. Variant interpretation: functional assays to the rescue. Am. J. Human Genet. 101, 315–325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brnich SE et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 12, 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartl DL & Clark AG Principles of Population Genetics 4th edn (Sinauer Associates, 1989). [Google Scholar]
- 67.Cassa CA et al. Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet. 49, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charlesworth B & Hill WG Selective effects of heterozygous protein-truncating variants. Nat. Genet. 51, 2 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mulder N et al. H3Africa: current perspectives. Pharmgenomics Pers. Med. 11, 59–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siepel A et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Glorot X, Bordes A & Bengio Y Deep sparse rectifier neural networks. In Proc. 14th International Conference on Artificial Intelligence and Statistics 315–323 (JMLR, 2011). [Google Scholar]
- 73.Ke G, He D & Liu T-Y Rethinking positional encoding in language pre-training. In 2021 International Conference on Learning Representations (ICLR, 2021). [Google Scholar]
- 74.Bateman A Uniprot: a universal hub of protein knowledge. Protein Sci. 28, 32–32 (2019). [Google Scholar]
- 75.Remmert M, Biegert A, Hauser A & Soding J HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Herrero J et al. Ensembl comparative genomics resources. Database 2016, bav096 (2016). [DOI] [PMC free article] [PubMed]
- 77.Klausen MS et al. NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins 87, 520–527 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Armean IM et al. Enhanced access to extensive phenotype and disease annotation of genes and genetic variation in Ensembl. Eur. J. Human Genet. 27, 1721–1721 (2019). [Google Scholar]
- 79.McLaren W et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ge R, Kakade SM, Kidambi R & Netrapalli P Rethinking learning rate schedules for stochastic optimization. In 2019 International Conference on Learning Representations (ICLR, 2018). [Google Scholar]
- 81.Zhang H & Shen Y ShenLab/gMVP: v1.0.0-alpha. Zenodo 10.5281/zenodo.7134878 (2022). [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pre-computed gMVP scores for all possible missense variants in canonical transcripts on human hg38 can be downloaded from https://www.dropbox.com/s/nce1jhg3i7jw1hx/gMVP.2021–02-28.csv.gz?dl=0. The training data of the main model were downloaded from http://www.discovehrshare.com/downloads (DiscovEHR), http://www.hgmd.cf.ac.uk/ac/index.php (HGMD), https://www.uniprot.org/docs/humpvar (UniProt) and https://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37/ (ClinVar). Other datasets supporting the findings of this study are available in the paper and the Supplementary Information.