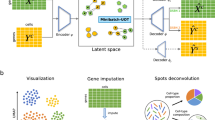
Abstract
Single-cell multimodal datasets have measured various characteristics of individual cells, enabling a deep understanding of cellular and molecular mechanisms. However, multimodal data generation remains costly and challenging, and missing modalities happen frequently. Recently, machine learning approaches have been developed for data imputation but typically require fully matched multimodalities to learn common latent embeddings that potentially lack modality specificity. To address these issues, we developed an open-source machine learning model, Joint Variational Autoencoders for multimodal Imputation and Embedding (JAMIE). JAMIE takes single-cell multimodal data that can have partially matched samples across modalities. Variational autoencoders learn the latent embeddings of each modality. Then, embeddings from matched samples across modalities are aggregated to identify joint cross-modal latent embeddings before reconstruction. To perform cross-modal imputation, the latent embeddings of one modality can be used with the decoder of the other modality. For interpretability, Shapley values are used to prioritize input features for cross-modal imputation and known sample labels. We applied JAMIE to both simulation data and emerging single-cell multimodal data including gene expression, chromatin accessibility, and electrophysiology in human and mouse brains. JAMIE significantly outperforms existing state-of-the-art methods in general and prioritized multimodal features for imputation, providing potentially novel mechanistic insights at cellular resolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MMD-MA simulation dataset can be downloaded from https://noble.gs.washington.edu/proj/mmd-ma/. Our simulation data may be downloaded from https://github.com/daifengwanglab/JAMIE. Processed Patch-seq gene expression and electrophysiological features for the mouse visual and motor cortices are available at https://github.com/daifengwanglab/scMNC. Raw Patch-seq datasets are available at refs. 2,31. Single-cell RNA-seq and ATAC-seq data on the developing human brain can be downloaded at https://github.com/GreenleafLab/brainchromatin/blob/main/links.txt under the heading ‘Multiome’. Single-cell RNA-seq and ATAC-seq of colon adenocarcinoma data can be found at https://github.com/wukevin/babel. Processed datasets for SNARE-seq adult mouse cortex data can be downloaded from https://scglue.readthedocs.io/en/latest/data.html.
Code availability
All code was implemented in Python using PyTorch, and the source code is publicly available at https://github.com/daifengwanglab/JAMIE33. Since Code Ocean provides an interactive platform for computational reproducibility34, we have also provided an interactive version of our code for reproducing results and figures35.
Change history
16 June 2023
In the version of this article initially published Daifeng Wang was not solely listed as the corresponding author, and the contact information has now been amended in the HTML and PDF versions of the article.
References
Cadwell, C. R. et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 34, 199–203 (2016).
Gouwens, N. W. et al. Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell 183, 935–953 (2020).
Trevino, A. E. et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069 (2021).
Nguyen, N.D., Huang, J. & Wang, D. A deep manifold-regularized learning model for improving phenotype prediction from multi-modal data. Nat. Comput. Sci. 2, 38–46 (2022).
Wu, K. E., Yost, K. E., Chang, H. Y. & Zou, J. BABEL enables cross-modality translation between multiomic profiles at single-cell resolution. Proc. Natl Acad. Sci. USA 118, e2023070118 (2021).
Zhang, R, Meng-Papaxanthos, L, Vert, J.-P. & Noble, W. S. In Research in Computational Molecular Biology (ed. Pe’er, I.) 20–35 (Springer International, 2022).
Cao, K., Bai, X., Hong Y. & Wan, L. Unsupervised topological alignment for single-cell multi-omics integration. Bioinformatics 36, i48–i56 (2020).
Liu, J., Huang, Y., Singh, R., Vert J.-P. & Noble, W. S. Jointly embedding multiple single-cell omics measurements. WABI. 143, 10:1–10:13 (2019).
Cao, Z.-J. & Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 40, 1458–1466 (2022).
Zhang, Z., Yang, C. & Zhang, X. scDART: integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol. 23, 139 (2022).
Khan, S. A. et al. scAEGAN: Unification of single-cell genomics data by adversarial learning of latent space correspondences. PLoS ONE 18, e0281315 (2023).
Zhu, J.-Y., Park, T., Isola P. & Efros, A. A. Unpaired image-to-image translation using cycle-consistent adversarial networks. ICCV (2017).
Gala, R. et al. Consistent cross-modal identification of cortical neurons with coupled autoencoders. Nat. Comput. Sci. 1, 120–127 (2021).
Tu, X., Cao, Z.-J., Xia, C.-R., Mostafavi, S., & Gao, G. Cross-Linked Unified Embedding for cross-modality representation learning. Adv. Neural Inf. Process. Syst. 35, 15942–15955 (2022).
Nguyen, N. D., Blaby, I. K. & Wang, D. ManiNetCluster: a novel manifold learning approach to reveal the functional links between gene networks. BMC Genomics 20, 1003 (2019).
Hotelling, H. Relations between two sets of variates. Biometrika 28, 321–377 (1936).
Gala, R. et al. A coupled autoencoder approach for multi-modal analysis of cell types. NeurIPS, 32, 9263-9272 (2019).
Lundberg, S. & Lee, S.-I. A Unified Approach to Interpreting Model Predictions. NeurIPS, 31, 4768-4777 (2017).
Johansen, N. & Quon, G. scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 20, 1–21 (2019).
Li, H., Zhang, Z., Squires, M., Chen, X. & Zhang, X. scMultiSim: simulation of multi-modality single cell data guided by cell–cell interactions and gene regulatory networks. Preprint at https://www.biorxiv.org/content/10.1101/2022.10.15.512320v3 (2022).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Quinn, L. A., Moore, G. E., Morgan, R. T. & Woods, L. K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 39, 4194–4924 (1979).
Shi, J., Cheng, C., Ma, J., Liew, C.-C. & Geng, X. Gene expression signature for detection of gastric cancer in peripheral blood. Oncol. Lett. 15, 9802–9810 (2018).
Bergdolt, L. & Dunaevsky, A. Brain changes in a maternal immune activation model of neurodevelopmental brain disorders. Prog. Neurobiol. 175, 1–19 (2019).
Harder, J. M. & Libby, R. T. BBC3 (PUMA) regulates developmental apoptosis but not axonal injury induced death in the retina. Mol. Neurodegener. 6, 1–10 (2011).
Song, Y.-H. et al. Somatostatin enhances visual processing and perception by suppressing excitatory inputs to parvalbumin-positive interneurons in V1. Sci. Adv. 6, eaaz0517 (2020).
Kingma, D. P. & Welling, M. Auto-Encoding Variational Bayes. ICLR (2014).
Doersch, C. Tutorial on variational autoencoders. Arxiv Tech Report https://arxiv.org/abs/1606.05908 (2016).
Bowman, S. R. et al. Generating sentences from a continuous space. Assoc. Comput. Linguist. 57, 6008–6019 (2015).
Cui, Z., Change, H., Shan, S. & Chen, X. Generalized unsupervised manifold alignment. Adv. Neural Inform. Process. Syst. 3, 2429–2437 (2014).
Scala, F. et al. Phenotypic variation of transcriptomic cell types in mouse motor cortex. Nature 598, 144–150 (2021).
McInnes, L., Healy J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. J. Open Source Softw. 3, 861 (2018).
Cohen Kalafut, N., Huang, X. & Wang, D. Joint variational autoencoders for multimodal imputation and embedding. Zenodo https://doi.org/10.5281/zenodo.7782362 (2023).
Clyburne-Sherin, A., Fei X. & Green, S. A. Computational reproducibility via containers in social psychology. Meta-Psychology 3, 892 (2019).
Cohen Kalafut, N., Huang X. & Wang, D. Joint variational autoencoders for multimodal imputation and embedding. Code Ocean https://doi.org/10.24433/CO.0507883.v1 (2023).
Acknowledgements
This work was supported by National Institutes of Health grants R21NS128761, R21NS127432 and R01AG067025 to D.W., P50HD105353 to Waisman Center, National Science Foundation Career Award 2144475 to D.W. and the start-up funding for D.W. from the Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. The funders had no role in study design, data collection and analysis, decision to publish or manuscript preparation.
Author information
Authors and Affiliations
Contributions
D.W. conceived and supervised the study. N.C.K. developed and implemented the methodology. X.H. and D.W. verified the methods. N.C.K. performed visualization and analysis. N.C.K, X.H. and D.W. edited and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Xiuwei Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Additional comparison table, supporting method hyperparameters, detailed dataset information and Figs. 1–16.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cohen Kalafut, N., Huang, X. & Wang, D. Joint variational autoencoders for multimodal imputation and embedding. Nat Mach Intell 5, 631–642 (2023). https://doi.org/10.1038/s42256-023-00663-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00663-z
This article is cited by
-
Unsupervised data imputation with multiple importance sampling variational autoencoders
Scientific Reports (2025)
-
Variational graph autoencoder for reconstructed transcriptomic data associated with NLRP3 mediated pyroptosis in periodontitis
Scientific Reports (2025)
-
TMO-Net: an explainable pretrained multi-omics model for multi-task learning in oncology
Genome Biology (2024)
-
scButterfly: a versatile single-cell cross-modality translation method via dual-aligned variational autoencoders
Nature Communications (2024)
-
scPair: Boosting single cell multimodal analysis by leveraging implicit feature selection and single cell atlases
Nature Communications (2024)