Abstract
The COVID-19 pandemic emphasized the importance of epidemic forecasting for decision makers in multiple domains, ranging from public health to the economy. Forecasting epidemic progression is a non-trivial task due to multiple confounding factors, such as human behaviour, pathogen dynamics and environmental conditions. However, the surge in research interest and initiatives from public health and funding agencies has fuelled the availability of new data sources that capture previously unobservable aspects of disease spread, paving the way for a spate of ‘data-centred’ computational solutions that show promise for enhancing our forecasting capabilities. Here we discuss various methodological and practical advances and introduce a conceptual framework to navigate through them. First we list relevant datasets, such as symptomatic online surveys, retail and commerce, mobility and genomics data. Next we consider methods, focusing on recent data-driven statistical and deep learning-based methods, as well as hybrid models that combine domain knowledge of mechanistic models with the flexibility of statistical approaches. We also discuss experiences and challenges that arise in the real-world deployment of these forecasting systems, including decision-making informed by forecasts. Finally, we highlight some challenges and open problems found across the forecasting pipeline to enable robust future pandemic preparedness.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Holmdahl, I. & Buckee, C. Wrong but useful—what COVID-19 epidemiologic models can and cannot tell us. N. Engl. J. Med. 383, 303–305 (2020).
Marathe, M. & Vullikanti, A. K. S. Computational epidemiology. Commun. ACM 56, 88–96 (2013).
Biggerstaff, M., Cauchemez, S., Reed, C., Gambhir, M. & Finelli, L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect. Dis. 14, 480 (2014).
Viboud, C. et al. The RAPIDD ebola forecasting challenge: synthesis and lessons learnt. Epidemics 22, 13–21 (2018).
Johansson, M. A., Apfeldorf, K. M., Dobson, S. & Devita, J. et al. An open challenge to advance probabilistic forecasting for dengue epidemics. Proc. Natl Acad. Sci. USA 116, 24268–24274 (2019).
Cramer, E. Y. et al. The United States COVID-19 Forecast Hub dataset. Sci. Data 9, 462 (2022).
Sherratt, K. et al. Predictive performance of multi-model ensemble forecasts of COVID-19 across European nations. eLife 12, e81916 (2023).
Aktay, A. et al. Google COVID-19 Community Mobility Reports (Google, accessed 15 May 2024); https://www.google.com/covid19/mobility
Astley, C. M. et al. Global monitoring of the impact of the COVID-19 pandemic through online surveys sampled from the Facebook user base. Proc. Natl Acad. Sci. USA 118, e2111455118 (2021).
Peccia, J. et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 38, 1164–1167 (2020).
Biggerstaff, M. et al. Coordinating the real-time use of global influenza activity data for better public health planning. Influenza Other Respir. Virus. 14, 105–110 (2020).
Butler, P., Ramakrishnan, N., Nsoesie, E. O. & Brownstein, J. S. Satellite imagery analysis: what can hospital parking lots tell us about a disease outbreak? IEEE Ann. Hist. Comput. 47, 94–97 (2014).
Miliou, I. et al. Predicting seasonal influenza using supermarket retail records. PLoS Comput. Biol. 17, e1009087 (2021).
Borchering, R. K. et al. Public health impact of the US Scenario Modeling Hub. Epidemics 44, 100705 (2023).
Biggerstaff, M. et al. Results from the centers for disease control and prevention's predict the 2013-2014 Influenza Season Challenge. BMC Infect. Dis. 16, 357 (2016).
Chakraborty, P. et al. What to know before forecasting the flu. PLoS Comput. Biol. 14, e1005964 (2018).
Cramer, E. Y. et al. Evaluation of individual and ensemble probabilistic forecasts of COVID-19 mortality in the United States. Proc. Natl Acad. Sci. USA 119, e2113561119 (2022).
Kandula, S. et al. Evaluation of mechanistic and statistical methods in forecasting influenza-like illness. J. R. Soc. Interf. 15, 20180174 (2018).
Kandula, S. & Shaman, J. Near-term forecasts of influenza-like illness: an evaluation of autoregressive time series approaches. Epidemics 27, 41–51 (2019).
Reich, N. G. et al. A collaborative multiyear, multimodel assessment of seasonal influenza forecasting in the United States. Proc. Natl Acad. Sci. USA 116, 3146–3154 (2019).
Bracher, J. On the multibin logarithmic score used in the FluSight competitions. Proc. Natl Acad. Sci. USA 116, 20809–20810 (2019).
Gneiting, T. & Raftery, A. E. Strictly proper scoring rules, prediction, and estimation. J. Am. Stat. Assoc. 102, 359–378 (2007).
Hethcote, H. W. The mathematics of infectious diseases. SIAM Rev. 42, 599–653 (2000).
Viboud, C. & Vespignani, A. The future of influenza forecasts. Proc. Natl Acad. Sci. USA 116, 2802–2804 (2019).
Polgreen, P. M., Chen, Y., Pennock, D. M., Nelson, F. D. & Weinstein, R. A. Using internet searches for influenza surveillance. Clin. Infect. Dis. 47, 1443–1448 (2008).
Ginsberg, J. et al. Detecting influenza epidemics using search engine query data. Nature 457, 1012–1014 (2009).
Culotta, A. Towards detecting influenza epidemics by analyzing Twitter messages. In Proc. First Workshop on Social Media Analytics 115–122 (ACM, 2010).
Yang, S., Santillana, M. & Kou, S. C. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc. Natl Acad. Sci. USA 112, 14473–14478 (2015).
Ning, S., Yang, S. & Kou, S. C. Accurate regional influenza epidemics tracking using Internet search data. Sci. Rep. 9, 5238 (2019).
Ray, E. L. & Reich, N. G. Prediction of infectious disease epidemics via weighted density ensembles. PLoS Comput. Biol. 14, e1005910 (2018).
Chakraborty, P. et al. Forecasting a moving target: ensemble models for ILI case count predictions. In Proc. 2014 SIAM International Conference on Data Mining (eds Zaki, M. et al.) 262–270 (SIAM, 2014).
Zou, B., Lampos, V. & Cox, I. Multi-task learning improves disease models from web search. In Proc. 2018 World Wide Web Conference 87–96 (IW3C2, 2018).
Matsubara, Y., Sakurai, Y., Van Panhuis, W. G. & Faloutsos, C. FUNNEL: automatic mining of spatially coevolving epidemics. In Proc. 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 105–114 (ACM, 2014).
Wang, Z. et al. Dynamic poisson autoregression for influenza-like-illness case count prediction. In Proc. 21st ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 1285–1294 (ACM, 2015).
Lamb, A., Paul, M. J. & Dredze, M. Separating fact from fear: tracking flu infections on Twitter. In Proc. 2013 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (eds Vanderwende, L. et al.) 789–795 (ACL, 2013).
Zou, B., Lampos, V. & Cox, I. Transfer learning for unsupervised influenza-like illness models from online search data. In Proc. 2019 World Wide Web Conference (eds Liu, L. & White, R.) 2505–2516 (ACM, 2019).
Paul, M. J. & Dredze, M. A model for mining public health topics from Twitter. Johns Hopkins University https://www.cs.jhu.edu/~mdredze/publications/2011.tech.twitter_health.pdf (2011).
Chen, L., Tozammel Hossain, K. S. M., Butler, P., Ramakrishnan, N. & Prakash, B. A. Flu gone viral: syndromic surveillance of flu on Twitter using temporal topic models. In 2014 IEEE International Conference on Data Mining 755–760 (IEEE, 2014).
Rekatsinas, T. et al. SourceSeer: forecasting rare disease outbreaks using multiple data sources. In Proc. 2015 SIAM International Conference on Data Mining (eds Venkatasubramanian, S. & Ye, J.) 379–387 (SIAM, 2015).
Brooks, L. C., Farrow, D. C., Hyun, S., Tibshirani, R. J. & Rosenfeld, R. Flexible modeling of epidemics with an empirical Bayes framework. PLoS Comput. Biol. 11, e1004382 (2015).
Ray, E. L., Sakrejda, K., Lauer, S. A., Johansson, M. A. & Reich, N. G. Infectious disease prediction with kernel conditional density estimation. Stat. Med. 36, 4908–4929 (2017).
Brooks, L. C., Farrow, D. C., Hyun, S., Tibshirani, R. J. & Rosenfeld, R. Nonmechanistic forecasts of seasonal influenza with iterative one-week-ahead distributions. PLoS Comput. Biol. 14, e1006134 (2018).
Zimmer, C. & Yaesoubi, R. Influenza forecasting framework based on Gaussian processes. In Proc. 37th International Conference on Machine Learning (eds Daumé, H. III & Singh, A.) 11671–11679 (PMLR, 2020).
Senanayake, R., O’Callaghan, S. & Ramos, F. Predicting spatio–temporal propagation of seasonal influenza using variational Gaussian process regression. In Proc. 30th AAAI Conference on Artificial Intelligence 3901–3907 (AAAI, 2016).
Volkova, S., Ayton, E., Porterfield, K. & Corley, C. D. Forecasting influenza-like illness dynamics for military populations using neural networks and social media. PLoS ONE 12, e0188941 (2017).
Ayyoubzadeh, S. M., Ayyoubzadeh, S. M., Zahedi, H., Ahmadi, M. & Kalhori, S. R. N. Predicting COVID-19 incidence through analysis of google trends data in Iran: data mining and deep learning pilot study. JMIR Publ. Health Surveill. 6, e18828 (2020).
Venna, S. R. et al. A novel data-driven model for real-time influenza forecasting. IEEE Access 7, 7691–7701 (2018).
Adhikari, B., Xu, X., Ramakrishnan, N. & Prakash, B. A. EpiDeep: exploiting embeddings for epidemic forecasting. In Proc. 25th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 577–586 (ACM, 2019).
Wang, L. et al. Examining deep learning models with multiple data sources for COVID-19 forecasting. In 2020 IEEE International Conference on Big Data (Big Data) 3846–3855 (IEEE, 2020).
Jin, X., Wang, Y.-X. & Yan, X. Inter-series attention model for COVID-19 forecasting. In Proc. 2021 SIAM International Conference on Data Mining (eds Demeniconi, C. et al.) 495–503 (SIAM, 2021).
Wu, Y., Yang, Y., Nishiura, H. & Saitoh, M. Deep learning for epidemiological predictions. In 41st International ACM SIGIR Conference on Research & Development in Information Retrieval 1085–1088 (ACM, 2018).
Deng, S., Wang, S., Rangwala, H., Wang, L. & Ning, Y. Cola-GNN: Cross-location attention based graph neural networks for long-term ILI prediction. In Proc. 29th ACM International Conference on Information & Knowledge Management 245–254 (ACM, 2020).
Roy, P. et al. Deep diffusion-based forecasting of COVID-19 by incorporating network-level mobility information. In Proc. 2021 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (eds Coscia, M. et al.) 168–175 (ACM, 2021).
Ibrahim, M. R. et al. Variational-LSTM autoencoder to forecast the spread of coronavirus across the globe. PLoS ONE 16, e0246120 (2021).
Ramchandani, A., Fan, C. & Mostafavi, A. DeepCOVIDNet: an interpretable deep learning model for predictive surveillance of COVID-19 using heterogeneous features and their interactions. IEEE Access 8, 159915–159930 (2020).
Rodríguez, A. et al. Steering a historical disease forecasting model under a pandemic: case of flu and COVID-19. In Proc. 35th AAAI Conference on Artificial Intelligence 4855–4863 (AAAI, 2021).
Panagopoulos, G., Nikolentzos, G. & Vazirgiannis, M. Transfer graph neural networks for pandemic forecasting. In Proc. 35th AAAI Conference on Artificial Intelligence 4838–4845 (AAAI, 2021).
Kamarthi, H., Kong, L., Rodríguez, A., Zhang, C. & Prakash, B. A. When in doubt: neural non-parametric uncertainty quantification for epidemic forecasting. In Proc. 35th Conference on Neural Information Processing Systems (eds Ranzato, M. et al.) 19796–19807 (NeurIPS, 2021).
Kamarthi, H., Kong, L., Rodríguez, A., Zhang, C. & Prakash, B. A. CAMul: calibrated and accurate multi-view time-series forecasting. In Proc. ACM Web Conference 2022 (eds Laforest, F. et al.) 3174–3185 (ACM, 2022).
Shaman, J. & Karspeck, A. Forecasting seasonal outbreaks of influenza. Proc. Natl Acad. Sci. USA 109, 20425–20430 (2012).
Kandula, S., Pei, S. & Shaman, J. Improved forecasts of influenza-associated hospitalization rates with Google Search Trends. J. R. Soc. Interf. 16, 20190080 (2019).
Pei, S. & Shaman, J. Aggregating forecasts of multiple respiratory pathogens supports more accurate forecasting of influenza-like illness. PLoS Comput. Biol. 16, e1008301 (2020).
Tabataba, F. S. et al. Epidemic forecasting framework combining agent-based models and smart beam particle filtering. In 2017 IEEE International Conference on Data Mining 1099–1104 (IEEE, 2017).
Zhang, Q. et al. Forecasting seasonal influenza fusing digital indicators and a mechanistic disease model. In Proc. 26th International Conference on World Wide Web 311–319 (ACM, 2017).
Wang, R., Maddix, D., Faloutsos, C., Wang, Y. & Yu, R. Bridging physics-based and data-driven modeling for learning dynamical systems. In Proc. 3rd Conference on Learning for Dynamics and Control (eds Jadbabaie, A. et al.) 385–398 (PMLR, 2021).
Arık, S. Ö. et al. A prospective evaluation of AI-augmented epidemiology to forecast COVID-19 in the USA and Japan. npj Digit. Med. 4, 1–18 (2021).
Qian, Z., Alaa, A. M. & van der Schaar, M. When and how to lift the lockdown? Global COVID-19 scenario analysis and policy assessment using compartmental Gaussian processes. In Proc. 34th Conference on Neural Information Processing Systems 10729–10740 (NeurIPS, 2020).
Chopra, A. et al. Differentiable agent-based epidemiology. In Proc. 2023 International Conference on Autonomous Agents and Multiagent Systems 1848–1857 (International Foundation for Autonomous Agents and Multiagent Systems, 2023).
Osthus, D., Gattiker, J., Priedhorsky, R. & Del Valle, S. Y. et al. Dynamic Bayesian influenza forecasting in the United States with hierarchical discrepancy (with discussion). Bayes. Anal. 14, 261–312 (2019).
Osthus, D. & Moran, K. R. Multiscale influenza forecasting. Nat. Commun. 12, 2991 (2021).
Wu, D. et al. DeepGLEAM: a hybrid mechanistic and deep learning model for COVID-19 forecasting. Preprint at https://arxiv.org/abs/2102.06684 (2021).
Kamarthi, H., Rodríguez, A. & Prakash, B. A. Back2Future: leveraging backfill dynamics for improving real-time predictions in future. In Proc. Tenth International Conference on Learning Representations (ICLR, 2022).
Wang, L., Chen, J. & Marathe, M. DEFSI: deep learning based epidemic forecasting with synthetic information. In Proc. 33rd AAAI Conference on Artificial Intelligence 9607–9612 (AAAI, 2019).
Rodríguez, A., Cui, J., Ramakrishnan, N., Adhikari, B. & Prakash, B. A. EINNs: epidemiologically-informed neural networks. In Proc. 37th AAAI Conference on Artificial Intelligence (eds Williams, B. et al.) 14453–14460 (AAAI, 2023).
Kargas, N. et al. STELAR: spatio-temporal tensor factorization with latent epidemiological regularization. In Proc. 35th AAAI Conference on Artificial Intelligence 4830–4837 (AAAI, 2021).
Recchia, G., Freeman, A. L. & Spiegelhalter, D. How well did experts and laypeople forecast the size of the COVID-19 pandemic? PLoS ONE 16, e0250935 (2021).
Shea, K. et al. Harnessing multiple models for outbreak management. Science 368, 577–579 (2020).
Polgreen, P. M., Nelson, F. D., Neumann, G. R. & Weinstein, R. A. Use of prediction markets to forecast infectious disease activity. Clin. Infect. Dis. 44, 272–279 (2007).
Farrow, D. C. et al. A human judgment approach to epidemiological forecasting. PLoS Comput. Biol. 13, e1005248 (2017).
McAndrew, T., Cambeiro, J. & Besiroglu, T. Aggregating human judgment probabilistic predictions of the safety, efficacy, and timing of a COVID-19 vaccine. Vaccine 40, 2331–2341 (2022).
Reich, N. G. et al. Accuracy of real-time multi-model ensemble forecasts for seasonal influenza in the US. PLoS Comput. Biol. 15, e1007486 (2019).
Adiga, A. et al. All models are useful: Bayesian ensembling for robust high resolution COVID-19 forecasting. In Proc. 27th ACM SIGKDD Conference on Knowledge Discovery and Data Mining 2505–2513 (ACM, 2021).
McAndrew, T. & Reich, N. G. Adaptively stacking ensembles for influenza forecasting. Stat. Med. 40, 6931–6952 (2021).
Kim, J.-S., Kavak, H., Züfle, A. & Anderson, T. COVID-19 ensemble models using representative clustering. SIGSPATIAL Special 12, 33–41 (2020).
Altieri, N. et al. Curating a COVID-19 data repository and forecasting county-level death counts in the United States. Harv. Data Sci. Rev. https://doi.org/10.1162/99608f92.1d4e0dae (2021).
Rodríguez, A. et al. DeepCOVID: an operational deep learning-driven framework for explainable real-time COVID-19 forecasting. In Proc. 35th AAAI Conference on Artificial Intelligence 15393–15400 (AAAI, 2021).
Ferguson, N. M. et al. Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID-19 Mortality and Healthcare Demand (Imperial College COVID-19 Response Team, 2020).
Reich, N., Tibshirani, R., Ray, E. & Rosenfeld, R. On the predictability of COVID-19. IIF Blog https://forecasters.org/blog/2021/09/28/on-the-predictability-of-covid-19 (2021).
Probert, W. J. et al. Real-time decision-making during emergency disease outbreaks. PLoS Comput. Biol. 14, e1006202 (2018).
Nikolopoulos, K., Punia, S., Schäfers, A., Tsinopoulos, C. & Vasilakis, C. Forecasting and planning during a pandemic: COVID-19 growth rates, supply chain disruptions, and governmental decisions. Euro. J. Oper. Res. 290, 99–115 (2021).
Atkins, B. D. et al. Anticipating future learning affects current control decisions: a comparison between passive and active adaptive management in an epidemiological setting. J. Theor. Biol. 506, 110380 (2020).
Shea, K., Tildesley, M. J., Runge, M. C., Fonnesbeck, C. J. & Ferrari, M. J. Adaptive management and the value of information: learning via intervention in epidemiology. PLoS Biol. 12, e1001970 (2014).
Mhasawade, V., Zhao, Y. & Chunara, R. Machine learning and algorithmic fairness in public and population health. Nat. Mach. Intell. 3, 659–666 (2021).
Zhang, C. et al. A survey on federated learning. Knowl. Based Syst. 216, 106775 (2021).
Reinhart, A. et al. An open repository of real-time COVID-19 indicators. Proc. Natl Acad. Sci. USA 118, e2111452118 (2021).
Scarpino, S. V. & Petri, G. On the predictability of infectious disease outbreaks. Nat. Commun. 10, 898 (2019).
Rosenkrantz, D. J. et al. Fundamental limitations on efficiently forecasting certain epidemic measures in network models. Proc. Natl Acad. Sci. USA 119, e2109228119 (2022).
Karniadakis, G. E. et al. Physics-informed machine learning. Nat. Rev. Phys. 3, 422–440 (2021).
Mastakouri, A. & Schölkopf, B. Causal analysis of COVID-19 Spread in Germany. Adv. Neur. Inf. Process. Syst. 33, 3153–3163 (2020).
Kraemer, M. U. et al. Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence. Science 373, 889–895 (2021).
Ray, E. L. et al. Ensemble forecasts of coronavirus disease 2019 (COVID-19) in the U.S. Preprint at medRxiv https://doi.org/10.1101/2020.08.19.20177493 (2020).
Riquelme, C. et al. Scaling vision with sparse mixture of experts. In Proc. 35th Conference on Neural Information Processing Systems (eds Ranzato, M. et al.) 8583–8595 (NeurIPS, 2021).
Angelini, G., De Angelis, L. & Singleton, C. Informational efficiency and behaviour within in-play prediction markets. Int. J. Forecast. 38, 282–299 (2022).
Lutz, C. S. et al. Applying infectious disease forecasting to public health: a path forward using influenza forecasting examples. BMC Public Health 19, 1659 (2019).
Pollett, S. et al. Recommended reporting items for epidemic forecasting and prediction research: the EPIFORGE 2020 guidelines. PLoS Med. 18, e1003793 (2021).
Gibson, G. C., Reich, N. G. & Sheldon, D. Real-time mechanistic Bayesian forecasts of COVID-19 mortality. Ann. Appl. Stat. 17, 1801–1819 (2023).
Wu, J. T., Leung, K. & Leung, G. M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 395, 689–697 (2020).
Edeling, W. et al. The impact of uncertainty on predictions of the CovidSim epidemiological code. Nat. Comput. Sci. 1, 128–135 (2021).
Balcan, D. et al. Multiscale mobility networks and the spatial spreading of infectious diseases. Proc. Natl Acad. Sci. USA 106, 21484–21489 (2009).
Pei, S., Kandula, S., Yang, W. & Shaman, J. Forecasting the spatial transmission of influenza in the United States. Proc. Natl Acad. Sci. USA 115, 2752–2757 (2018).
Chang, S. et al. Mobility network models of COVID-19 explain inequities and inform reopening. Nature 589, 82–87 (2021).
Gopalakrishnan, V. et al. Globally local: hyper-local modeling for accurate forecast of COVID-19. Epidemics 37, 100510 (2021).
Geng, X. et al. A kernel-modulated SIR model for COVID-19 contagious spread from county to continent. Proc. Natl Acad. Sci. USA 118, e2023321118 (2021).
Santillana, M., Nsoesie, E. O., Mekaru, S. R., Scales, D. & Brownstein, J. S. Using clinicians’ search query data to monitor influenza epidemics. Clin. Infect. Dis. 59, 1446–1450 (2014).
Soebiyanto, R. P., Adimi, F. & Kiang, R. K. Modeling and predicting seasonal influenza transmission in warm regions using climatological parameters. PLoS ONE 5, e9450 (2010).
Paul, M. & Dredze, M. You are what you tweet: analyzing Twitter for public health. In Proc. 5th International AAAI Conference on Weblogs and Social Media 265–272 (AAAI, 2011).
Ghamizi, S. et al. Data-driven simulation and optimization for COVID-19 exit strategies. In Proc. 26th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 3434–3442 (ACM, 2020).
Osthus, D. Fast and accurate influenza forecasting in the United States with Inferno. PLoS Comput. Biol. 18, e1008651 (2022).
Nadella, P., Swaminathan, A. & Subramanian, S. Forecasting efforts from prior epidemics and COVID-19 predictions. Euro. J. Epidemiol. 35, 727–729 (2020).
Hemming, V., Burgman, M. A., Hanea, A. M., McBride, M. F. & Wintle, B. C. A practical guide to structured expert elicitation using the IDEA protocol. Methods Ecol. Evol. 9, 169–180 (2018).
Viboud, C., Boëlle, P.-Y., Carrat, F., Valleron, A.-J. & Flahault, A. Prediction of the spread of influenza epidemics by the method of analogues. Am. J. Epidemiol. 158, 996–1006 (2003).
Acknowledgements
This work was supported in part by the National Science Foundation (grant numbers Expeditions CCF-1918770, CAREER IIS-2028586, RAPID IIS-2027862, Medium IIS-1955883, Medium IIS-2106961, CCF-2115126 and PIPP CCF-2200269), the CDC MInD programme, the ORNL, faculty research awards from Facebook and funds/computing resources from Georgia Tech.
Author information
Authors and Affiliations
Contributions
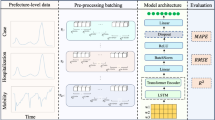
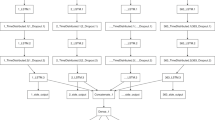
A.R., H.K. and B.A.P. contributed to the conceptualization of the manuscript. All authors contributed to gathering, analysing and interpreting the literature. P.A., J.H., M.P. and S.S. contributed to the development of Figs. 1 and 2. A.R., H.K. and B.A.P. contributed to the writing of all sections.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Sen Pei and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and text.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rodríguez, A., Kamarthi, H., Agarwal, P. et al. Machine learning for data-centric epidemic forecasting. Nat Mach Intell 6, 1122–1131 (2024). https://doi.org/10.1038/s42256-024-00895-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-024-00895-7
This article is cited by
-
Identifying and forecasting importation and asymptomatic spreaders of multi-drug resistant organisms in hospital settings
npj Digital Medicine (2025)
-
Integrating artificial intelligence with mechanistic epidemiological modeling: a scoping review of opportunities and challenges
Nature Communications (2025)