Abstract
Objective
The HIV epidemic remains a significant public health issue in the United States. HIV risk prediction models could be beneficial for reducing HIV transmission by helping clinicians identify patients at high risk for infection and refer them for testing. This would facilitate initiation on treatment for those unaware of their status and pre-exposure prophylaxis for those uninfected but at high risk. Existing HIV risk prediction algorithms rely on manual construction of features and are limited in their application across diverse electronic health record systems. Furthermore, the accuracy of these models in predicting HIV in females has thus far been limited.
Materials and methods
We devised a pipeline for automatic construction of prediction models based on automatic feature engineering to predict HIV risk and tested our pipeline on a local electronic health records system and a national claims data. We also compared the performance of general models to female-specific models.
Results
Our models obtain similarly good performance on both health record datasets despite difference in represented populations and data availability (AUC = 0.87). Furthermore, our general models obtain good performance on females but are also improved by constructing female-specific models (AUC between 0.81 and 0.86 across datasets).
Discussion and conclusions
We demonstrated that flexible construction of prediction models performs well on HIV risk prediction across diverse health records systems and perform as well in predicting HIV risk in females, making deployment of such models into existing health care systems tangible.
Keywords: HIV, risk prediction, electronic health records, HIV prevention, predictive modeling
Background and significance
Despite major improvements in HIV diagnosis and treatment over recent decades, the HIV epidemic is a continuing problem in the United States. The Centers for Disease Control and Prevention (CDC) estimate there were approximately 1.2 million people with HIV (PWH) living in the United States in 2019.1 This includes an estimated 13% who are undiagnosed and remain unaware of their infection. Testing and diagnosis of these individuals is of paramount importance for reaching the target goal of ending the HIV epidemic in the United States by 2030,2 as these individuals account for 40% of new HIV infections.
Another important tool for reaching these public health goals involves the use of pre-exposure prophylaxis (PrEP) for persons at high risk of HIV infection, introduced in 2012 for adults and 2018 for patients ≤18 years old.3,4 However, despite the availability and effectiveness of PrEP, uptake has been slow with only 23% of people eligible for the treatment having a current prescription in 2019.5 Several barriers to increasing PrEP use have been identified.6–8 Chief among these is providers not regularly screening for indications for PrEP because a very small proportion (<1%) of people in the United States are estimated to need this intervention.
There is a clear need for tools to help providers identify individuals at high risk for HIV infection to facilitate initiation of conversations about sexual history, HIV testing, and PrEP. Machine learning-based risk prediction models derived from electronic health record (EHR) data are an example of such tools that have been explored in recent years for HIV risk prediction.9,10 Krakower et al,11 Marcus et al,12 and Ahlström et al13 each developed HIV risk prediction models using EHR data from a multidisciplinary outpatient practice in Boston, MA, Kaiser Permanente Northern California system, and a national medical record data in Denmark, respectively.
While these risk models performed well, they have 2 major limitations. First, these models were all developed using manually selected and engineered features relying on clinical expertise such as “number of positive tests for gonorrhea or chlamydia in the previous 2 years.” Engineering such features is time-consuming and may not generalize well across EHR systems. The second major limitation of previously published HIV risk models is while they perform well in predominantly male populations, they perform very poorly when identifying females at risk of HIV infection. The model developed by Marcus et al12 flagged 46% of males with incident HIV, however, it flagged none of the females, while the other studies did not evaluate their models on females at all. While the rate of new HIV infections has been declining in males over the last 5 years, the rate in females has remained stable, demonstrating a need for better prevention implementation strategies in this population.1 Furthermore, only 10% of females with an indication for PrEP were prescribed the medication in 2019.14 A risk model that can reliably identify females at high risk of HIV infection as well as males would be an invaluable tool for clinicians to improve the uptake of PrEP in this population.
Objective
In order to address the challenge of HIV prevention, we identified 2 critical tasks that can significantly reduce new HIV cases: (1) improving identification of undiagnosed PWH to connect them to effective treatments, protecting both their health and reducing the risk of transmission; and (2) identifying high risk individuals who can benefit from PrEP. We address these 2 challenges, while undertaking the 2 aforementioned limitations of previous models in this study. We developed a pipeline for creating risk models for HIV infection based on automatic feature engineering and demonstrated that this pipeline can be automatically applied across different types of EHR and claims data. We tested our method on national claims data and local EHR data and show that female-specific risk models improve on the performance of the general models in this population.
Methods
Data
Two clinical databases were used for the development and evaluation of the models. Optum’s de-identified Clinformatics Data Mart Database (CDM) is a national de-identified database derived from administrative health claims from members of large commercial and Medicare Advantage health plans. In addition to administrative data, the database contains data on prescribed medications and laboratory tests, including test results. The database has records for approximately 68 million patients, from all 50 US states and spans over 13 years from January 1, 2007, through June 30, 2020. The UT Physicians (UTP) clinical data warehouse stores EHR data from the UTPs outpatient network based in Houston, TX. This database contains records for approximately 4 million patients from 2005 through 2021. The use of these databases in the study was approved by the UTHealth Committee for the Protection of Human Subjects.
Cohort selection and preparation
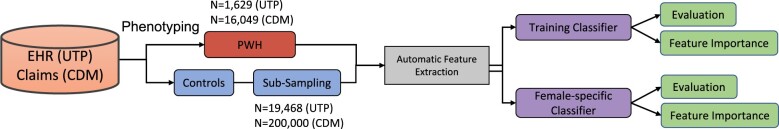
Development of HIV risk prediction models depends on accurate identification of PWH in the data. Identifying new HIV diagnoses (ie, identifying the first HIV diagnosis of a patient) in the EHR is difficult and has been based on specific sequence of HIV diagnostic tests, codes, and measurement of HIV-1 plasma RNA levels in previous studies.15 In our study, we instead identified all PWH in different stages based on our previously published phenotyping algorithm.16 People with HIV were identified in both CDM and UTP databases (Figure 1) using this algorithm. Briefly, patients with a positive HIV confirmatory test, detectable HIV viral load greater than 1000 copies/mL, or prescription for HIV antiretroviral medications specifically for treatment rather than prevention of HIV were considered to have a diagnosis of HIV. Inclusion criteria involved patients at least 13 years of age based on the CDC recommended minimal age for universal HIV screening.17 Patients who are not PWH were considered the control pool for this study. An index date was then assigned to PWH as the date of the earliest evidence for HIV infection in the data, that is, the minimum date of first ICD code for HIV, first positive HIV screening test, first positive HIV confirmatory test, first detectable viral load, or first antiretroviral prescription date. For the controls, the index date was the date of their last encounter in the database. We excluded patients if their length of clinical history prior to the index date was less than 1 year. By requiring at least 1 year of history prior to the first evidence of HIV to be included in the study, we reduce the risk of including prior HIV diagnoses into our analysis data as controls.
Figure 1.
Schematic of cohort derivation and modeling process. Abbreviations: CDM: Clinformatics Data Mart Database; EHR: electronic health record; PWH: people with HIV; UTP: UT Physicians.
Feature extraction and data preparation
We extracted 4 types of features from the data, including demographics (race, sex, age at index date, marital status [available in UTP only]), and diagnoses (ICD 9-CM and ICD 10 diagnosis codes), prescribed medications, and laboratory data, all 3 types from a 2-year window prior to index date. For ICD codes, we used only top-level codes, that is, the first 3 characters describing the disease category. Medications were coded in generic names and represented as dichotomous features (ie, has the patient used the medication within the observation period or not). We disregarded the doses or packaging information. The 2 datasets vary in the included medications. The CDM contains only medications that had both prescription and dispensing dates while UTP included prescription records as well as recorded medications (medications a patient reports taking but were not prescribed in the system). We constructed features from laboratory tests using both the numeric value of the laboratory result and a dichotomous feature indicating the presence of the laboratory test. To handle the longitudinal values of lab tests, the numeric results per patient were aggregated using the average, minimum and maximum values across the 2-year window prior to index date followed by scaling each feature using z-score. Missing numeric data were imputed using k-nearest neighbor (KNN, k = 3) imputation. We removed sparse features that were present in less than 5% of patients in both the PWH and the control group. Categorical features were represented using one-hot encoding, where we create a new column (dummy variable) for each unique value in the category and fill each dummy variable with ones if the sample has this category and zero otherwise.
Model optimization and training
Due to the large numeric imbalance between PWH and control populations, we selected 10 random subsamples of control patients, each including approximately the same number of controls as there were PWH. We tested 6 classifiers: logistic regression, Least Absolute Shrinkage and Selection Operator (LASSO) and Ridge penalized logistic regression, random forest,18 and 2 types of gradient boosting decision tree algorithms, XGBoost19 and CatBoost.20 Since the boosted tree models can handle missing data, these algorithms were also tested on data without imputation. Model hyperparameters were tuned in a nested 5-fold cross-validation via Bayesian optimization21 using the BayesianSearchCV class, part of the Python scikit-optimize library.22 All models were developed using Python version 3.9 and scikit-learn version 1.1.1.
Evaluation
Evaluation metrics included recall (sensitivity), precision (positive predictive value; PPV), F1, and AUC, averaged across the 5-folds for each subsample and further averaged across 10 subsamples of control patients to obtain the final value of these metrics for each model. We compared the performance of the 6 different classifiers on the full datasets to determine which classifier performed the best at this task. We then compared the performance of the top model trained only on females to the performance of the top model trained on the full dataset and tested in female patients only. We further evaluated the contribution of the 20 top-performing features using Shapley Additive Explanations (SHAP values23).
We ensured the low risk of bias and clinical utility by passing the checklists of the Prediction model Risk Of Bias ASsessment Tool (PROBAST) and the CHecklist for critical Appraisal and data extraction for systematic Reviews of prediction Modelling Studies (CHARMS)24 using the template proposed in Fernandez-Felix et al.25
Results
Demographics of PWH and controls in UTP and CDM datasets
The UTP dataset included 1629 PWH and 19 468 randomly sampled control patients meeting the criteria for inclusion in the analyses with the year of index date ranging between 2005 and 2021. For training the models, we created 10 subsamples from the controls and ran the models 10 times (Methods). Comparisons of the demographic characteristics between PWH and controls in the dataset are summarized in Table 1. Compared to controls, PWH were younger (43.3 ± 15.6 and 48.7 ± 20.7 for PWH and controls, respectively), more often male (59% for PWH vs 39% for controls), more often single (43% PWH vs 24% controls were single while 15% PWH were married vs 30% of controls), and more often Black (48% for PWH vs 19% for controls), following trends seen among PWH in local and national surveillance data.5,26
Table 1.
Comparison of demographic characteristics between PWH and control patients for UTP (N = 21 097) and CDM (N = 216 049) datasets.
| UTP |
CDM |
|||||
|---|---|---|---|---|---|---|
| Control | PWH | P | Control | PWH | P | |
| n | 19 468 | 1629 | 200 000 | 16 049 | ||
| Sex (%) | ||||||
| M | 7616 (39.1) | 966 (59.3) | <.001 | 89 148 (44.6) | 12 375 (77.1) | <.001 |
| F | 11 872 (61.0) | 663 (40.7) | <.001 | 110 841 (55.4) | 3673 (22.9) | <.001 |
| Race/Ethnicity (%) | ||||||
| White | 8922 (45.8) | 461 (28.3) | <.001 | 126 807 (63.4) | 8424 (52.5) | <.001 |
| Black | 3631 (18.7) | 776 (47.6) | <.001 | 18 759 (9.4) | 3568 (22.2) | <.001 |
| Hispanic | 1310 (6.7) | 111 (6.8) | .936 | 20 885 (10.4) | 2614 (16.3) | <.001 |
| Other | 4593 (23.6) | 258 (15.8) | <.001 | n/a | n/a | |
| Unknown | 1056 (5.4) | 29 (1.8) | <.001 | 25 137 (12.6) | 838 (5.2) | <.001 |
| Age at index date (mean (SD)) | 48.70 (20.74) | 43.31 (15.58) | <.001 | 51.75 (19.28) | 38.50 (13.12) | <.001 |
| Age at index date (%) | <.001 | <.001 | ||||
| 13-24 | 3046 (15.6) | 152 (9.6) | 17 389 (8.7) | 2211 (13.8) | ||
| 25-34 | 2774 (14.2) | 285 (18.0) | 28 046 (14.0) | 4832 (30.2) | ||
| 35-44 | 2782 (14.3) | 332 (21.0) | 30 915 (15.5) | 3910 (24.4) | ||
| 45-54 | 2722 (14.0) | 396 (25.1) | 32 218 (16.1) | 3051 (19.0) | ||
| 55+ | 8144 (41.8) | 414 (26.2) | 91 432 (45.7) | 2016 (12.6) | ||
| Marital status (%) | ||||||
| Married | 5824 (29.9) | 240 (14.7) | <.001 | n/a | n/a | |
| Single | 4709 (24.2) | 707 (43.4) | <.001 | n/a | n/a | |
| Other | 1325 (6.8) | 120 (7.4) | .34 | n/a | n/a | |
| Unknown | 7610 (39.1) | 562 (34.5) | <.001 | n/a | n/a | |
| Year of index date (%) | <.001 | <.001 | ||||
| 2005 | 49 (0.3) | 33 (2.0) | n/a | n/a | ||
| 2006 | 118 (0.6) | 102 (6.3) | n/a | n/a | ||
| 2007 | 132 (0.7) | 106 (6.5) | n/a | n/a | ||
| 2008 | 138 (0.7) | 73 (4.5) | 7377 (3.7) | 732 (4.6) | ||
| 2009 | 150 (0.8) | 108 (6.6) | 9581 (4.8) | 989 (6.2) | ||
| 2010 | 215 (1.1) | 76 (4.7) | 9028 (4.5) | 1042 (6.5) | ||
| 2011 | 241 (1.2) | 69 (4.2) | 9248 (4.6) | 995 (6.2) | ||
| 2012 | 261 (1.3) | 57 (3.5) | 9143 (4.6) | 1030 (6.4) | ||
| 2013 | 399 (2.0) | 91 (5.6) | 12 702 (6.4) | 1100 (6.9) | ||
| 2014 | 658 (3.4) | 80 (4.9) | 106 48 (5.3) | 1076 (6.7) | ||
| 2015 | 603 (3.1) | 107 (6.6) | 10 130 (5.1) | 1358 (8.5) | ||
| 2016 | 867 (4.5) | 89 (5.5) | 11 807 (5.9) | 1605 (10.0) | ||
| 2017 | 1288 (6.6) | 131 (8.0) | 12 699 (6.3) | 1571 (9.8) | ||
| 2018 | 1953 (10.0) | 144 (8.8) | 15 165 (7.6) | 1798 (11.2) | ||
| 2019 | 2760 (14.2) | 151 (9.3) | 24 618 (12.3) | 1956 (12.2) | ||
| 2020 | 3890 (20.0) | 138 (8.5) | 57 854 (28.9) | 797 (5.0) | ||
| 2021 | 5746 (29.5) | 74 (4.5) | n/a | n/a | ||
Abbreviations: CDM: Clinformatics Data Mart Database; n/a, not applicable; PWH: people with HIV; UTP: UT Physicians
The CDM dataset included 16 049 PWH and 200 000 randomly sampled controls patients who met the criteria for inclusion in the analyses. The year of index date for these patients ranged from 2008 to 2020 (Table 1). Similar to the UTP data, PWH were younger and more often male in the CDM data compared to the control patients (77% vs 45% male, respectively). While the overall population in CDM includes more White patients than the population in the UTP dataset, PWH in the CDM dataset were more likely to be Black than controls.
Model training and evaluation in UTP and CDM datasets
We evaluated 6 machine learning algorithms on the UTP and CDM datasets (Methods, Table 2). XGBoost models performed the best on both UTP and CDM data (precision = 0.76 and 0.75, recall = 0.73 and 0.75, F1 = 0.74 and 0.75, and AUC = 0.85 and 0.86 on UTP and CDM data, respectively, Table 2). XGBoost models were also trained with the unimputed version of the data (Methods), displaying slightly improved performance, with a precision of 0.77 and 0.76, recall of 0.75 and 0.76, and the same F1 of 0.76 and AUC of 0.87 in both UTP and CDM data, respectively (Table 2). We thus chose the XGBoost model trained on unimputed data as the best performing model to use for further analyses.
Table 2.
Model evaluation results for UTP and CDM datasets (all metrics given as mean (SD)).
| Indicator plus numeric variables for laboratory tests—UTP | ||||
|---|---|---|---|---|
| Model | Precision | Recall | F 1 | AUC |
| Logistic regression | 0.73 (0.006) | 0.74 (0.007) | 0.73 (0.006) | 0.83 (0.005) |
| LASSO (L1) | 0.74 (0.006) | 0.74 (0.007) | 0.74 (0.006) | 0.84 (0.004) |
| Ridge (L2) | 0.75 (0.007) | 0.74 (0.008) | 0.74 (0.007) | 0.84 (0.004) |
| Random Forest | 0.73 (0.009) | 0.68 (0.008) | 0.70 (0.008) | 0.81 (0.005) |
| XGBoost—imputed data | 0.76 (0.009) | 0.73 (0.012) | 0.74 (0.009) | 0.85 (0.008) |
| XGBoost—unimputed data | 0.77 (0.009) | 0.75 (0.008) | 0.76 (0.008) | 0.87 (0.005) |
| CATBoost—imputed data | 0.75 (0.005) | 0.71 (0.006) | 0.73 (0.004) | 0.84 (0.005) |
| CATBoost—unimputed data | 0.77 (0.009) | 0.75 (0.006) | 0.76 (0.006) | 0.86 (0.004) |
| Indicator plus numeric variables for laboratory tests—CDM | ||||
|---|---|---|---|---|
| Model | Precision | Recall | F 1 | AUC |
| Logistic regression | 0.73 (0.001) | 0.75 (0.002) | 0.74 (0.002) | 0.85 (0.001) |
| LASSO (L1) | 0.73 (0.002) | 0.75 (0.002) | 0.74 (0.002) | 0.85 (0.001) |
| Ridge (L2) | 0.73 (0.002) | 0.75 (0.002) | 0.74 (0.002 | 0.85 (0.001) |
| Random Forest | 0.75 (0.002) | 0.72 (0.002) | 0.73 (0.002) | 0.84 (0.001) |
| XGBoost—imputed data | 0.75 (0.002) | 0.75 (0.002) | 0.75 (0.002) | 0.86 (0.001) |
| XGBoost—unimputed data | 0.76 (0.003) | 0.76 (0.002) | 0.76 (0.002) | 0.87 (0.001) |
| CATBoost—imputed data | 0.75 (0.002) | 0.75 (0.002) | 0.75 (0.002) | 0.86 (0.001) |
| CATBoost—unimputed data | 0.76 (0.003) | 0.75 (0.003) | 0.76 (0.002) | 0.87 (0.001) |
Abbreviations: CDM: Clinformatics Data Mart Database; UTP: UT Physicians. Maximal AUC is in marked in bold.
Owing to the difference in the way index date was defined for cases and controls, controls tended to have a later index date. This created a potential bias in ICD-9 versus ICD-10 usage, since ICD-10 was officially implemented in October 2015.27 To verify that our models are not affected by this potential bias, we selected a subset of patients (cases and controls) whose index date occurred from 2017 on, allowing for the 2-year window to include the time after the transition to ICD-10 codes. We observed similar performance on this cohort using our selected algorithm of XGBoost (Table S1).
Evaluation of best performing model in females
We evaluated the top performing model in Table 3. The model trained on the full dataset had a slightly higher AUC than the model trained specifically on female patients in both datasets (0.86 vs 0.85 for UTP and 0.81 vs 0.80 for CDM). However, we found that the model trained specifically on female patients had an improved precision and recall. While in UTP data the improvement was minor (precision of 0.75 vs 0.74 in the fully trained model and recall of 0.66 vs 0.64), the improvement was more substantial in the CDM data (precision: 0.70 vs 0.67 in the fully trained model; and recall: 0.72 vs 0.36). Similar to the full dataset, we also tested our method only patients with index date after 2017, obtaining comparable results in UTP and better performance in the CDM (Table S1).
Table 3.
Model evaluation results for UTP and CDM in female-only datasets (all metrics given as mean (SD)).
| XGBoost—UTP | ||||
|---|---|---|---|---|
| Model | Precision | Recall | F 1 | AUC |
| Overall model—full dataset | 0.77 (0.009) | 0.75 (0.008) | 0.76 (0.008) | 0.87 (0.005) |
| Overall model—female-only | 0.74 (0.01) | 0.64 (0.01) | 0.69 (0.01) | 0.86 (0.01) |
| Female-specific model | 0.75 (0.014) | 0.66 (0.012) | 0.70 (0.012) | 0.85 (0.009) |
| XGBoost—CDM | ||||
|---|---|---|---|---|
| Model | Precision | Recall | F 1 | AUC |
| Overall model—full dataset | 0.76 (0.003) | 0.76 (0.002) | 0.76 (0.002) | 0.87 (0.001) |
| Overall model—female-only | 0.67 (0.006) | 0.36 (0.005) | 0.47 (0.006) | 0.81 (1e−16) |
| Female-specific model | 0.70 (0.004) | 0.72 (0.008) | 0.71 (0.003) | 0.80 (0.003) |
Abbreviations: CDM: Clinformatics Data Mart Database; UTP: UT Physicians.
Top features of best performing models in overall and female-specific data
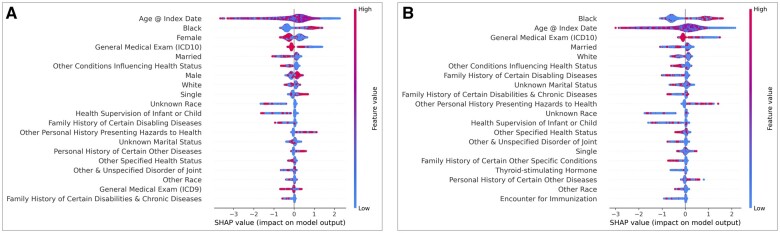
The top 20 features contributing to the UTP model can be seen in Figure 2A and a list the features their associated codes and data type (diagnosis, medication, laboratory test) can be found in Table S1. Overall, the top features consisted of demographic variables, features suggesting healthcare utilization (eg, ICD codes for general medical exam), and personal or family history of disease or exposure to health hazards (eg, smoking). Shapley Additive Explanation values associated with these features (x-axis, Figure 3) were aligned with the demographic traits described in the previous section, that is, that patients predicted to have HIV were more often young, Black, male, and single. They also tended to have ICD codes for personal history of hazards to health and certain other diseases and lacked ICD codes suggesting regular interaction with the healthcare system.
Figure 2.
Top 20 features in UTP data ranked by feature importance for overall model (A) and female-specific model (B). Abbreviations: SHAP: Shapley Additive Explanation; UTP: UT Physicians.
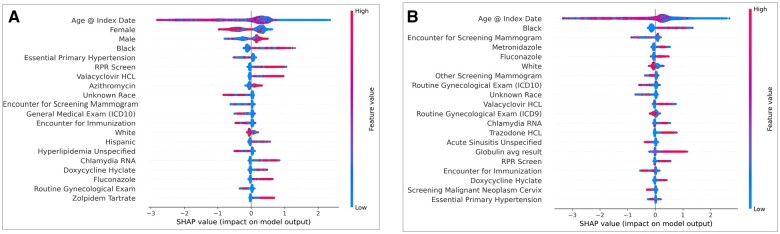
Figure 3.
Top 20 features in CDM data ranked by feature importance for overall model (A) and female-specific model (B). Abbreviation: SHAP: Shapley Additive Explanation.
Similar to the overall model, the top 20 features for the female-specific XGBoost model in the UTP data demonstrated similar trends in the types of features contributing the most information to the model (Figure 2B).
In the CDM data, the top features in the overall XGBoost model included demographic features (PWH are more often young, male, and Black or Hispanic) and ICD codes reflecting regular healthcare utilization (PWH had fewer), similar to the UTP data (Figure 3A). A list of the top feature with their associated codes and data type can be seen in Table S2. However, the top features in the CDM data also included laboratory tests screening for sexually transmitted infections (STIs; RPR Screen, Chlamydia RNA) as well as medications commonly used to treat STIs and other infections, such as valacyclovir, azithromycin, fluconazole, and doxycycline. Finally, the top features seen in the female-specific XGBoost model in the CDM data were similar to the model trained on all the data, albeit different ranking (Figure 3B).
Discussion
In this study, we developed a pipeline to generate risk prediction models for HIV infection. Our pipeline uses automatic feature engineering, overcoming previous reliance on manually defined features, and we show that they are generalizable across different types of data (EHR and claims data) and perform well in different populations (population local to greater Houston, TX vs population sampled from all over the United States), with our top performing model achieving an AUC of 0.87 in both UTP and CDM datasets.
Models for HIV risk prediction developed using EHR data have been previously published. A LASSO penalized logistic regression model was developed using hand-selected features derived from EHR data in Boston, MA by Krakower et al.11 This model had a cross validated AUC of 0.86. In a companion study, Marcus et al12 developed a similar LASSO penalized logistic regression model using EHR data from the Kaiser Permanente Northern California system which had a cross validated AUC of 0.84. Finally, Ahlström et al13 developed a ridge penalized logistic regression HIV risk prediction model using a national registry in Denmark which included demographic variables and information on past medical history. This model achieved an AUC of 0.88 on a validation dataset. In comparison, our top performing model, an XGBoost model, performed similarly to these previously published models with a cross validated AUC of 0.87 in both the UTP and CDM datasets. The similar result with our approach in the absence of curating features is a major advantage of our approach. In addition, the 2 datasets we used to develop and evaluate our models are different in many respects. The CDM is nationally derived and UTP is from the Houston region. The CDM data are de-identified and derived from insurance claims data that have been augmented with medication and laboratory result data. In contrast, the UTP data is a fully identified dataset derived from the EHR system of the UTPs outpatient network. While both datasets contain mainly privately insured patients, they have different demographic distributions, particularly among PWH (Table 1). That we were able to develop models that equally well identify patients with HIV in both datasets demonstrates the portability of our modeling process. Our results also suggest that building system-specific or population-specific models using features automatically generated from the data could improve the translatability of risk prediction models between health systems, as they often differ from one system to another, and features that might be present in data from one system may not be available in another.
Assessing HIV risk in females is challenging for several reasons. The primary risk factor for HIV infection in men is having sex with other men (MSM), which can be queried relatively easily in a medical history. By default, cis-gender women cannot be MSM. Therefore, determining a female’s HIV risk requires the clinician taking a detailed sexual history as part of a medical visit, however this is often not done.28,29 Even if a sexual history is taken, the female patient might not be aware of the HIV risk of her partners. In a data-driven approach to identifying high-risk females, the population of PWH is heavily skewed male (approximately 80%)1 meaning the number of training examples of females with HIV is small by comparison. Furthermore, the strongest risk factors for HIV (ie, MSM) are negative for females by default thus drowning out any potential risk they might have for HIV infection and making the model more likely to erroneously flag them as being low risk. We demonstrate that our models perform well in females (AUC 0.86 in UTP data and 0.81 in CDM data for model trained on the full cohort) but training it on a subset of female patients further improves the quality of HIV risk prediction in this population by substantially improving the precision and recall of the model. This is an important finding as previous models developed for prediction of HIV risk have either not been tested in females or have performed poorly in this population. For example, Marcus et al12 tested the ability of their model to identify PWH stratified by sex, and found that their model identified 46% of males with HIV but none of the females with HIV. We estimate that the primary reason for this bias in previous models is that their features were manually selected. Features that were pre-selected to address the overall performance might have been biased by the majority of PWH being males. Better targeting of HIV testing and PrEP to females is a major unmet clinical need.
Interestingly, in the UTP data, we saw similar features among the top 20 for both the model trained on the entire dataset and the model trained only on females, with some small shifts in the order of the top features between the 2 models. Conversely, in the CDM female-specific model, features related to gynecological health had higher importance than in the general model. Additionally, medications such as metronidazole and fluconazole were included that are often used to treat gynecologic concerns like pelvic inflammatory disease, bacterial vaginosis, and candidiasis. Average globulin level was a top feature in the female-specific model in CDM but not in the general model, with higher average globulin level being associated with a prediction of HIV diagnosis. It has been shown that higher globulin levels are seen in PWH as compared to people who are uninfected,30 however, to our knowledge, no difference with respect to sex has been previously reported. It is possible that the importance of this feature is obscured in the full dataset due to the dominance of male-specific factors.
Finally, the bulk of the top features in the UTP data suggest regular contact with the healthcare system, such as ICD 9 or ICD 10 codes for “general medical exam”, “other conditions influencing health status”, “health supervision of infant or child”, and “encounter for immunization.” Generally, people flagged as having HIV by the models lacked these features, suggesting that individuals at the highest risk of HIV infection have less interaction or access to regular healthcare. This is borne out by data demonstrating that populations hardest hit by the HIV epidemic tend to be of lower socioeconomic status (SES), lack health insurance, and have poor access to care.31,32
One limitation of our study is that the population included in both datasets used in this study consists of primarily privately insured patients. HIV infection disproportionately affects those who are generally of lower SES and thus are less likely to have private health insurance.32 Further tests will be needed to evaluate our models on populations without health insurance. Another limitation of our approach is that we train a new model for each dataset. While a unified model would be clinically desirable, such a model might not be practical due to differences in data availability across different health record datasets. Finally, when training the algorithm on years where ICD-9 codes were used (prior to 2015), a careful design needs to be added to make sure there is not imbalance between cases and controls. In our datasets we observed minor changes when constraining to patients after 2017, with small decrease in the UTP dataset, possibly resulting from lower samples size and increase in performance in the CDM.
Our results demonstrate that a model tailored to the data could be done automatically and is expected to have better performance than a manually curated model that attempts to bridge differences in clinical data by focusing only on the common denominator between these datasets. We suggest that HIV prevention could significantly benefit from HIV risk stratification, triaging individuals based on a computed HIV risk. This HIV risk can facilitate targeted screening and, as appropriate, offering of PrEP. While the CDC recommends testing every individual aged 13 to 64 at least once, in practice less than 40% of people in the United States have ever been tested for HIV, according to a CDC report.33 Furthermore, a single test might not be sufficient to ensure detection in high-risk individuals. HIV risk stratification can support providers in identifying those patients for whom more frequent screening would be appropriate. We thus propose that running our algorithm on a health system regularly may have the potential to identify changes in the risk of a potential patient and promote clinicians to suggest HIV screening.
Conclusion
In conclusion, we developed and evaluated a modeling strategy for predicting risk of HIV infection based on automatic feature engineering. Our strategy performs well across 2 different types of health data. Additionally, our models outperform previously published models when identifying females at risk for HIV infection and perform similarly to models trained specifically on female patients. Although outside of the scope of this study, we aim to follow up on patients admitted to the UTP system to prospectively validate which percentage of high-risk patients has contracted HIV in the coming years. Implementation of these models in clinical settings has the potential to help providers identify patients at high risk of HIV infection and provide testing and prevention interventions to those who need them most.
Supplementary Material
Contributor Information
Sarah B May, Center for Precision Health, McWilliams School of Biomedical Informatics, University of Texas Health Science Center at Houston, Houston, TX 77030, United States; Dan L Duncan Institute for Clinical and Translational Research, Baylor College of Medicine, Houston, TX 77030, United States.
Thomas P Giordano, Section of Infectious Diseases, Department of Medicine, Baylor College of Medicine, Houston, TX 77030, United States; Center for Innovations in Quality, Effectiveness and Safety, Michael E. DeBakey VA Medical Center, Houston, TX 77021, United States.
Assaf Gottlieb, Center for Precision Health, McWilliams School of Biomedical Informatics, University of Texas Health Science Center at Houston, Houston, TX 77030, United States.
Author contributions
S.B.M. conceived the study, performed the analyses, and drafted the manuscript. T.P.G. provided clinical guidance. S.B.M., T.P.G., and A.G. wrote the manuscript.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
S.B.M. was supported by the NLM Training Program in Biomedical Informatics and Data Science (T15LM007093). T.P.G. was supported by the MD Anderson Foundation Chair at Baylor College of Medicine and by the Texas Developmental Center for AIDS Research (P30AI161943).
Conflicts of interest
None declared.
Data availability
The data underlying this article (UTP) cannot be shared publicly because it contains fully identified health record data from patients in the UT Physicians outpatient network. A de-identified version of the data will be shared on reasonable request to the corresponding author. The data underlying this article (CDM) were provided by Optum under license/by permission. Data will be shared on request to the corresponding author with permission of Optum.
References
- 1.Volume 32 | HIV Surveillance | Reports | Resource Library | HIV/AIDS | CDC. Diagnoses of HIV Infection in the United States and Dependent Areas 2019, 2021. Accessed August 7, 2023. https://www.cdc.gov/hiv/library/reports/hiv-surveillance/vol-32/index.html
- 2. Fauci AS, Redfield RR, Sigounas G, Weahkee MD, Giroir BP.. Ending the HIV epidemic: a plan for the United States. JAMA. 2019;321(9):844-845. 10.1001/jama.2019.1343 [DOI] [PubMed] [Google Scholar]
- 3.Item of Interest: FDA approves PrEP therapy for adolescents at risk of HIV. Accessed November 6, 2023. https://www.nichd.nih.gov/newsroom/news/051618-PrEP
- 4.July HIVg, Published. FDA approves first drug for reducing the risk of sexually acquired HIV infection. HIV.gov, 2012. Accessed August 7, 2023. https://www.nichd.nih.gov/newsroom/news/051618-PrEP
- 5.National Profile | Volume 26 Number 2 | HIV Surveillance | Reports | Resource Library | HIV/AIDS | CDC. Monitoring Selected National HIV Prevention and Care Objectives by Using HIV Surveillance Data United States and 6 Dependent Areas, 2019: National Profile; 2022. Accessed August 7, 2023. https://www.cdc.gov/hiv/library/reports/hiv-surveillance/vol-26-no-2/content/national-profile.html
- 6. Calabrese SK, Tekeste M, Mayer KH, et al. Considering stigma in the provision of HIV pre-exposure prophylaxis: reflections from current prescribers. AIDS Patient Care STDS. 2019;33(2):79-88. 10.1089/apc.2018.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krakower D, Ware N, Mitty JA, Maloney K, Mayer KH.. HIV providers' perceived barriers and facilitators to implementing pre-exposure prophylaxis in care settings: a qualitative study. AIDS Behav. 2014;18(9):1712-1721. 10.1007/s10461-014-0839-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pleuhs B, Quinn KG, Walsh JL, Petroll AE, John SA.. Health care provider barriers to HIV pre-exposure prophylaxis in the United States: a systematic review. AIDS Patient Care STDS. 2020;34(3):111-123. 10.1089/apc.2019.0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marcus JL, Sewell WC, Balzer LB, Krakower DS.. Artificial intelligence and machine learning for HIV prevention: emerging approaches to ending the epidemic. Curr HIV/AIDS Rep. 2020;17(3):171-179. 10.1007/s11904-020-00490-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiang Y, Du J, Fujimoto K, Li F, Schneider J, Tao C.. Application of artificial intelligence and machine learning for HIV prevention interventions. Lancet HIV. 2022;9(1):e54-e62. 10.1016/S2352-3018(21)00247-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krakower DS, Gruber S, Hsu K, et al. Development and validation of an automated HIV prediction algorithm to identify candidates for pre-exposure prophylaxis: a modelling study. Lancet HIV. 2019;6(10):e696-e704. 10.1016/S2352-3018(19)30139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcus JL, Hurley LB, Krakower DS, Alexeeff S, Silverberg MJ, Volk JE.. Use of electronic health record data and machine learning to identify candidates for HIV pre-exposure prophylaxis: a modelling study. Lancet HIV. 2019;6(10):e688-e695. 10.1016/S2352-3018(19)30137-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahlström MG, Ronit A, Omland LH, Vedel S, Obel N.. Algorithmic prediction of HIV status using nation-wide electronic registry data. EClinicalMedicine. 2019;17:100203. 10.1016/j.eclinm.2019.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. CDC. HIV and Women. Centers for Disease Control and Prevention; 2022.
- 15. Goetz MB, Hoang T, Kan VL, Rimland D, Rodriguez-Barradas M.. Development and validation of an algorithm to identify patients newly diagnosed with HIV infection from electronic health records. AIDS Res Hum Retroviruses. 2014;30(7):626-633. 10.1089/AID.2013.0287 [DOI] [PubMed] [Google Scholar]
- 16. May SB, Giordano TP, Gottlieb A.. A phenotyping algorithm to identify people with HIV in electronic health record data (HIV-Phen): development and evaluation study. JMIR Form Res. 2021;5(11):e28620. 10.2196/28620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Centers for Disease Control and Prevention, Association of Public Health Laboratories. Laboratory Testing for the Diagnosis of HIV Infection: Updated Recommendations. 2014. Accessed November 6, 2023. https://stacks.cdc.gov/view/cdc/23447
- 18. Breiman L. Random forests. Mach Learn. 2001;45(1):5-32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 19. Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; August 13, 2016:785–794. [Google Scholar]
- 20. Dorogush AV, Ershov V, Gulin A. CatBoost: gradient boosting with categorical features support. arXiv preprint arXiv:1810.11363. 2018.
- 21. Frazier PI. A tutorial on Bayesian optimization. arXiv preprint arXiv:1807.02811. 2018.
- 22.scikit-optimize: sequential model-based optimization in Python–scikit-optimize 0.8.1 documentation. Accessed August 7, 2023. https://scikit-optimize.github.io/stable/
- 23. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30.
- 24. Moons KG, de Groot JA, Bouwmeester W, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11(10):e1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernandez-Felix BM, López-Alcalde J, Roqué M, Muriel A, Zamora J.. CHARMS and PROBAST at your fingertips: a template for data extraction and risk of bias assessment in systematic reviews of predictive models. BMC Med Res Methodol. 2023;23(1):44-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.HIV in the Houston Area. The 2019 Houston Area Integrated Epidemiologic Profile for HIV Prevention and Care Services Planning. Accessed November 6, 2023. https://www.houstonhealth.org/media/2886/download
- 27. Hirsch J, Nicola G, McGinty G, et al. ICD-10: history and context. AJNR Am J Neuroradiol. 2016;37(4):596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrow RY, Ahmed F, Bolan GA, Workowski KA.. Recommendations for providing quality sexually transmitted diseases clinical services, 2020. MMWR Recomm Rep. 2020;68(5):1-20. 10.15585/mmwr.rr6805a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wimberly YH, Hogben M, Moore-Ruffin J, Moore SE, Fry-Johnson Y.. Sexual history-taking among primary care physicians. J Natl Med Assoc. 2006;98(12):1924-1929. [PMC free article] [PubMed] [Google Scholar]
- 30. Patil R, Raghuwanshi U.. Serum protein, albumin, globulin levels, and A/G ratio in HIV positive patients. Biomed Pharmacol J. 2015;2(2):321-325. [Google Scholar]
- 31.Economically Disadvantaged | HIV by Group | HIV/AIDS | CDC; 2022. Accessed November 6, 2023. https://www.cdc.gov/hiv/group/poverty.html
- 32. Pellowski JA, Kalichman SC, Matthews KA, Adler N.. A pandemic of the poor: social disadvantage and the U.S. HIV epidemic. Am Psychol. 2013;68(4):197-209. 10.1037/a0032694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.NCHHSTP Communications Center - National Center for HIV/AIDS, Viral Hepatitis, STD and TB Communications Center, Centers for Disease Control and Prevention. Most Americans have never had an HIV test, new data show [Press release]. Accessed November 6, 2023. https://www.hiv.gov/blog/most-americans-have-never-had-hiv-test-new-data-show/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article (UTP) cannot be shared publicly because it contains fully identified health record data from patients in the UT Physicians outpatient network. A de-identified version of the data will be shared on reasonable request to the corresponding author. The data underlying this article (CDM) were provided by Optum under license/by permission. Data will be shared on request to the corresponding author with permission of Optum.