Abstract
The data representation as well as naming conventions used in commercial screen files by different companies make the automated analysis of crystallization experiments difficult and time-consuming. In order to reduce the human effort required to deal with this problem, we present an approach for computationally matching elements of two schemas using linguistic schema matching methods and then transform the input screen format to another format with naming defined by the user. This approach is tested on a number of commercial screens from different companies and the results of the experiments showed an overall accuracy of 97% on schema matching which is significantly better than the other two matchers we tested. Our tool enables mapping a screen file in one format to another format preferred by the expert using their preferred chemical names.
Keywords: schema matching, data integration, protein crystallization screen, consistent naming
I. Introduction
Protein crystallization is the process of forming protein crystals to determine the structure of a protein by using techniques such as X-ray crystallography for biological as well as industrial applications. Protein crystallization screening is the process of determining factors suitable for the formation of large protein crystals. The factors typically include types of reagents, molecular concentration, types of salts, pH value of buffers, temperature, etc [1], [2]. Many commercial companies have developed screen kits with a list of combinations of chemicals which could lead to successful crystalline conditions.
The commercial screen files obtained from different sources (companies) have inconsistent representations. The inconsistencies are due to i) the schema, that refers to the organization of data, and ii) variations in naming of the same chemical. One common way of storing screen files is spreadsheets. The columns or headers in a spreadsheet are an indicator of the schema although the developer may not have considered a specific schema ahead of time. The first problem, schema matching, is to match different headers from screen files. The second problem, consistent naming, is to represent the same chemical with different representations in a uniform way. Protein crystallization screening analysis and visualization tools such as Experimental Design (AED) [1], [3], GenScreen (a genetic algorithm) [4], [5], and Visual X2 [6], require a specific input screen file format for analyzing results of protein crystallization trial experiments. PickScreens [7] asks users to format the files for screen comparisons. Consistent naming is an important issue for identifying novel screens as addressed in the C6 web tool [8]. The differences between various commercial screens make it difficult to develop a tool that can effectively take input from them and thus leads to the need of a proper schema matching tool [9]. The goal in this paper is to map commercial screens to a format preferred by an expert while having consistent chemical names chosen by the expert.
Schema matching is the process of identifying semantic correspondences, also called matches, between the elements of two schemas by the use of their structural and syntactic pattern. Data integration involves combining data from several disparate sources and provide a unified view of the data. Data migration, a sub-area of data integration, is the process of transferring data from one system to another while changing the storage, database, or application [10]. In our application, we migrate the data from input to output database with different schemas using schema matching and data migration techniques but we will use the general terms data integration, transformation, and migration interchangeably in this paper. We refer to the screens of commercial screens as input schemas and the schemas to be used by analysis tools as output schemas.
Consistent naming of chemicals for protein crystallography has been discussed as an issue [11]. A chemical may be represented differently across multiple sources (using their common names, chemical formulas or scientific names). For example, ‘magnesium chloride’, ‘MgCl2’ and ‘magnesium dichloride’ represent the same chemical. These inconsistencies should be removed in order to avoid confusion and maintain uniformity and understandability in systems.
In our research, we use an individual schema-based, element-level, linguistic matcher to produce 1:1 mappings between the input and output schema rather than 1:n and m:n mappings. We propose a two step solution based on having a common intermediate schema (or global schema [12]) that acts as a bridge between any form of the commercial screen’s schema and the output schema. After matching schemas, our system analyzes chemical names, finds their International Union of Pure and Applied Chemistry (IUPAC)1 representations if available, use the IUPAC name as a reference name or a key of the chemical, and then lets the expert decide upon the preferred name for that chemical. After storing various representations of a chemical in the database, the system generates the output files with the expert’s preferred names.
Our contributions can be summarized as follows:
header (attribute) detection which may include the headers of multiple-rows, composite as well as split headers,
1:1 mapping (rather than 1:n or m:n mapping) between attributes with high accuracy thus significantly minimizing effort for corrections by the expert,
resolving the same chemical properties (attributes) of different chemical groups,
resolving mapping among enumerated chemical groups,
distinguishing chemical groups and chemical properties,
consistent naming of chemicals across screens based on expert preferences while allowing experts to use their naming convention, and
mapping a commercial screen to a desired screen format (even mapping between commercial screens).
The rest of the paper is organized as follows. Section II provides the background and related works in schema matching and integration. Section III presents the details of our method, similarity metrics used in the linguistic matching, and the process of data integration. Section IV analyzes the results of the experiments. Section V summarizes the paper.
II. Background and Related Works
A. Commercial Screens Dataset
Typically, protein crystallization involves a solution that includes three types of reagents: a precipitant, a buffer-controlling pH, and an additive (e.g., salt). A condition or a cocktail can be seen as a combination of these reagents at various concentrations [13]. A screen is a set of cocktails. Various commercial screens are prepared by different companies which contain a number of cocktail combinations which can lead to the growth of crystals. The commercial screens are usually provided as a spreadsheet file which contains the reagents under different headers. A reagent may have its own properties like name, concentration value, its unit, and pH value.
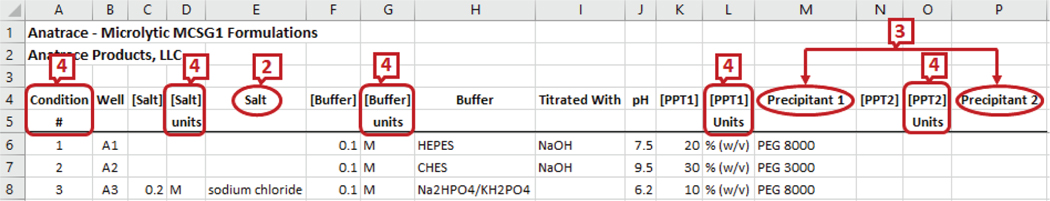
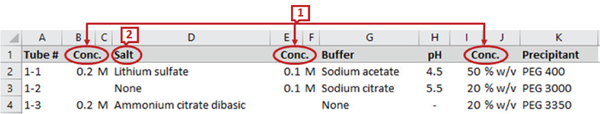
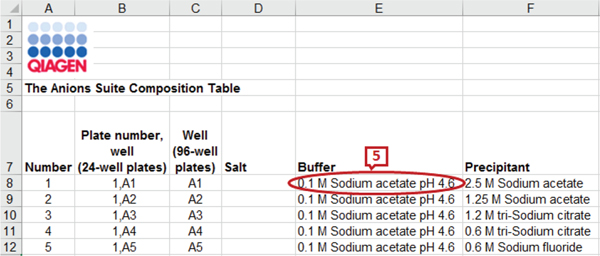
The commercial screen files from different companies have a different set of headers describing the reagents and their properties. For example, Fig. 1 and Fig. 2 show the snapshots of the headers and a few data rows of two screen files from Anatrace2 and Molecular Dimensions3 respectively. The marked labels in the figures are described in Section II-C. We can see from the figures that the number of reagents and their names in the screens are different. Some screen files even combine all the reagent properties under a single header. For example, the Anions Composition screen from Qiagen4 in Fig. 3 has concentration value, unit, and chemical name combined as a single attribute.
Fig. 1.
Snapshot of Anatrace Microlytic MCSG1 screen file
Fig. 2.
Snapshot of MD1–37 JCSG screen file
Fig. 3.
Snapshot of Qiagen’s Anions Suite screen file
B. Related Works on Schema Matching
A number of research studies have been done in the field of schema matching and integration. Our work was influenced by the linguistic techniques for schema matching used in SASMINT by Unal and Afsarmanesh, 2006 [14]. It makes effective use of NLP techniques and proposes the weighted usage of several syntactic and semantic similarity metrics. Cupid [15] is a hybrid approach which uses both linguistic and structural matching techniques based on the names, data types, constraints and schema structure to generate mappings between schema elements. Similarity flooding [16] is a graph matching algorithm which first transforms the two schemas into graphs and matches the strings of nodes between the two schemas. It also presents the overall measure for the human effort required to correct the results.
COMA [17] and COMA++ [18] are composite matchers designed for matching schema and ontology. COMA provides a large spectrum of individual matchers, in particular, a novel approach reusing previous match operations, and several mechanisms to combine the results of matcher executions. Harmony [19] is a schema matching tool that includes adding linguistic processing of textual documentation to conventional schema match techniques, learning from the input of a human in the loop, and GUI support for removing clutter and iterative development. FlexMatcher is a schema matching package in Python developed by the BigGorilla team [20] which uses a number of machine learning techniques to train a schema matcher using the information from the schemas and/or available instances. It then uses the trained matcher to make a prediction for matching the columns of the new schema to the mediated schema.
C. Limitations of Existing Systems
Although multiple tools are available for the purpose of schema matching and integration, several problems occur when dealing with a flat schema structure as in Microsoft (MS) Excel files that prevent the application of existing schema matchers to protein crystallization screens. Some information is repeated or aggregated in a flattened schema to make the instances identical [21]. Most of the tools require the schema to be defined in some defined formats like SQL, XML, etc. Hence they do not support repeated attribute names while protein crystallization screens may have columns with the same name describing different entities (e.g., ‘conc’ marked as 1 in Fig. 2). The header names are not always descriptive, for example, ‘salt’ (marked as 2 in Fig. 1 and 2) can match with any of ‘salt_name’, ‘salt_unit’ or ‘salt_concentration’ in another schema. There can be multivalued attributes of an entity in a screen schema which leads to enumeration of attributes (marked as 3 in Fig. 1). If there is only one precipitant in the target schema, it can match with any of the two precipitants in this schema. There may be multiple rows of headers in a spreadsheet and they may not necessarily start from the first row (marked as 4 in Fig. 1). Existing systems, if able to process spreadsheets, assume the first row as a header row. Composite attributes, having multiple property attributes, can be split into individual attributes or combined into a single attribute (e.g., label 5 in Fig. 3 shows the properties concentration, unit, name, and pH value in a single column). Also note that we do not use instance-based matching as the same chemical may appear under different headers or columns).
Furthermore, different screens use different representations for a specific property of a reagent which leads to the need of a domain-specific dictionary to map these representations. The previous algorithms are also unsuccessful in handling the cases where a source schema element has the same similarity score with multiple elements of target schema resulting in 1:n or n:1 mappings (e.g., salt could match with salt_name, salt_concentration, etc.). Since our purpose is the transformation of data from one schema to another, we want a 1:1 mapping between the source and target schema. 1:n, n:1, or m:n mappings may generate many unnecessary matches, and experts may rather do the matches by themselves rather than eliminating and correcting matches. Aggravating the problem, the commercial screens are not based on a well-designed schema and usually favor ease of use and readability. Hence, achieving 1:1 mapping is a critical and challenging task for achieving favorability by experts.
Although the standard naming of chemicals and reagents used in protein crystallography is an acknowledged problem [11], there are also no agreed standards for commercial crystallization screening software data representation and exchange. Newman et al. [8] propose using chemical classes for ambiguous chemicals (e.g., TRIS class, HEPES class). To the best of our knowledge, schema matching has not been applied for screen files. In this paper, we present our method while describing challenges faced and how we overcome those challenges.
Another issue is that of the buffer preparation. One can titrate the buffering species in its free acid or base form with the named counter ion, by adjusting the pH with the salt form of the buffer, or by titrating the salt form of the buffer to the final pH with the free acid or base form. Unless the vendors listings state differently we assume that the description 0.1M Buffer-Counter ion means that the solution is 0.1M in the buffering species, and that the pH is adjusted by titration with the counter ion.
III. Methodology
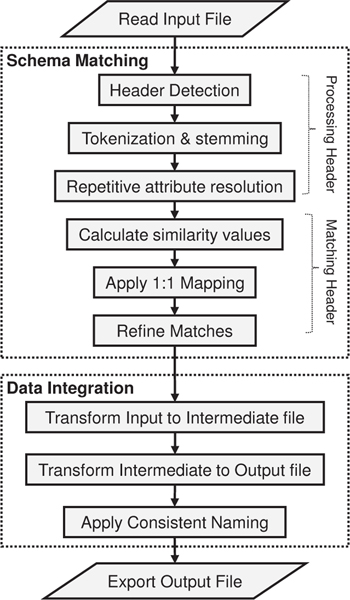
The primary goal of our research is to find the mappings between two schemas represented as column headers in MS Excel files so that data from the source file can be transformed to the target file under appropriate target schema headers. There are two main stages of our system: schema matching and data integration as shown in the flow diagram in Fig. 4. The schema matching stage matches the source schema with a pre-defined intermediate schema, which acts as a helper medium between the source and target schemas, and the intermediate schema with the target schema. The data integration stage uses these matches to transform the input screen to output screen also producing consistent names of chemicals.
Fig. 4.
Flow diagram of the system
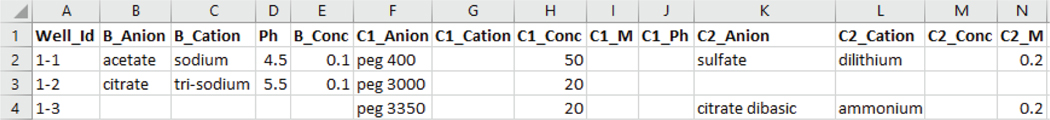
Since different commercial screens have different sets of headers for their screen files, we have come up with an intermediate schema in which the header names are more descriptive and can be matched with most of the screen files intuitively. The intermediate schema supports a buffer reagent, 3 precipitants and 2 salts. Each reagent has three properties: name, concentration value and its unit. The buffer reagent has an extra pH value property. The attributes of the intermediate schema are as follows: ‘well_id’, ‘buffer_name’, ‘buffer_conc’, ‘buffer_unit’, ‘ph’, ‘salt_name1’, ‘salt_conc1’, ‘salt_unit1’, ‘salt_name2’, ‘salt_conc2’, ‘salt_unit2’, ‘salt_name3’, ‘salt_conc3’, ‘salt_unit3’, ‘precipitant_name1’, ‘precipitant_conc1’, ‘precipitant_unit1’, ‘precipitant_name2’, ‘precipitant_conc2’, ‘precipitant_unit2’ [9]. An example of intermediate file produced by the system for MD1–37 JCSG screen file is shown in Fig. 5.
Fig. 5.
Snapshot of intermediate file for JCSG screen file
Instead of matching the input schema to the output schema directly, we make use of this intermediate schema to find input-to-intermediate and output-to-intermediate mappings. The use of the intermediate schema also adds flexibility to the mapping process between a number of input and output screens. For example, if there are m input screens and n output screens, the use of intermediate schema reduces the number of mappings from (m ∗ n) to (m + n). Moreover, this also enables mapping commercial screens between each other.
In the following subsections, we describe schema matching and data integration with consistent naming.
A. Processing Header for Schema Matching
In this research, we have used linguistic matching measures to identify similarities between schema element pairs. Linguistic or language-based matchers use the name and other textual elements of the schema to find semantically or syntactically similar elements. Here we have used the name of the header elements and their order in the input file to find related elements assuming that related attributes are likely to appear close to each other. Before applying linguistic measures, the schema elements from both the schemas were pre-processed through the following steps: header detection, tokenization & stemming of headers, and resolving repretitive attributes.
Header detection:
Most of the screens have only one header row which is the first row in the MS Excel file. But there are exceptions where additional information, for example, company name, screen information, etc. are provided in first a few rows and the actual headers only begin after several rows (e.g., Fig. 1). We consider the header row to be the row(s) just above the first occurrence of a numeric cell if it has a sufficient number of elements, i.e., more than two-third of the number of columns in the MS Excel file. This is however based on the assumption that the data rows contain at least one numeric value which is true for the screens seen so far. If the row just above the numeric cell does not have sufficient number of elements, there is a possibility that the file has multi-row headers. So, the two rows above the data row are concatenated in such cases to form the headers or attributes.
Tokenization & stemming:
After header detection, each header value is split into tokens based on white-spaces, underscore, and digits. Then white-spaces, empty tokens, and underscores are removed from the tokens. For example, the header ‘salt1 units’ would give ‘salt’, ‘1’ and ‘units’ as tokens. Stemming is applied to all tokens to change plural forms to singular (e.g., units to unit) and past tense verbs to root forms.
Repetitive attribute resolution:
The above-mentioned pre-processing steps are sufficient to help find the syntactic similarities in element names of screen files like Anatrace - Microlytic MCSG1 Formulations (Fig. 1) as they contain descriptive names like ‘salt conc’ and ‘ppt units’. Some other formats such as MD1–37 JCSG (Fig. 2) have repetition of headers or attribute names like ‘conc’ and ‘units’ which cannot be distinguished as ‘salt conc’, ‘buffer conc’ or ‘ppt conc’. For example, in Fig. 2, the header, ‘Conc.’ appears three times. In the schema, objects may share the same properties (attributes) leading to the repetition of attribute names. For example, both salt and precipitant have concentration values. The attributes related to the concentration value could be repeated for both salt and precipitant. To solve this problem, we prioritize some of the attributes as main attributes, for example, ‘salt’, ‘buffer’ and ‘precipitant’. The attributes such as concentration and unit are considered as property attributes and could be repeated. Then whenever tokens for property attributes such as ‘conc’ or ‘unit’ appear alone, previous token lists are searched for the appearance of main attributes and the search is stopped if either a main attribute is found or the beginning of the list is reached. If the beginning of the list is reached without finding any main attributes, search is continued in the next token lists. When a main attribute is found, it is appended to the token list. Some screens have multiple occurrences of main attributes and they are distinguished by a digit suffix, e.g., ‘salt1’ and ‘salt2’. If the tokens for the property attribute do not contain a digit but tokens for the related main attribute do, the digit value is also appended to the property token list along with the main attribute.
B. Matching Headers using Syntactic Similarity
Syntactical name matching computes the similarity solely based on comparing the name strings [18]. Our linguistic matcher uses approximate string matching techniques on the names of schema elements based on some similarity measures to find syntactically related element pairs.
Similarity Measures for Matching:
A single similarity measure is not effective in finding matches for all kinds of strings. Hence, we have used the following three similarity measures with different weights suited for our application: Levenshtein (Edit distance), Monge Elkan distance, and Term Frequency * Inverse Document Frequency (TF*IDF).
Levenshtein (Edit distance) is a measure of similarity between two strings based on the edit distance, i.e., the number of edit operations like insertions, deletions, and substitutions required to transform one string to another [22]. Before applying this measure, the tokens group of a header are concatenated to form a single string. Levenshtein similarity between two strings s and t is calculated as shown in (1) [14]:
| (1) |
where editDistance(s,t) is the minimal edit distance between s and t, and |s| and |t| are the lengths of strings s and t respectively.
Given two strings A and B with |A| and |B| as their respective number of tokens, the Monge-Elkan similarity is computed as in (2) [23]. Here we use Levenshtein similarity as the internal similarity measure sim’(a,b).
| (2) |
TF*IDF measure [24] assigns weights to terms. Term Frequency (TF) measures the number of times the token appears in a document (token group or a header element) and Inverse Document Frequency (IDF) measures how important the token is based on the number of documents (token groups) it appears on. TF*IDF similarity is calculated by computing similarity between individual tokens and then aggregating them based on their TF*IDF weights [25].
We tested the similarity measures individually with 6 screens: The Classics II Suite Composition, Anatrace – Microlytic MCSG1 Formulations, JBScreen Classic 1, MD1–02 Structure Screen, MD1–37 JCSG-plus, and MD1–104 The BCS Screen. We averaged the values of the evaluation measures across all 6 screens. Table I shows the results of the average performance of these 3 similarity measures. We use precision (P = TP/(TP + FP)), recall (R = TP/(TP + FN)) F1-measure (F1 = 2 ∗ P ∗ R/(P + R)) [24], and overall 8–20 ( measure, where TP is the number of true positives, FP is the number of false positives and FN is the number of false negatives. Overall measure [16] takes into account the amount of effort needed to add intended matches which have not been discovered (false negatives) and to remove the incorrect matches that have been proposed (false positives). Unlike F1-measure, overall is designed specifically for the purpose of assessing match quality. We can see that the Levenshtein similarity metric has the best performance on average followed by Monge Elkan and TF*IDF. Since we want to minimize the user effort in the matching process, we give more preference to the results of the similarity measures in terms of overall. We had also used Jaccard similarity [26] but it was not included due to its lower performance than other similarity measures. Hence, we use higher weights for Levenshtein and Monge Elkan measures and lower weight for the TF*IDF similarity measure while calculating the similarity matrix for our match algorithm:
| (3) |
where simLev, simME and simTFIDF are the similarity matrices formed using Levenshtein, Monge-Elkan and TF*IDF measures respectively. We also tried different combinations of the weights of these measures on trial and error basis and selected the combination which showed the best results.
TABLE I.
Average performance of 3 similarity measures
| Similarity Measures | Precision | Recall | F-measure | Overall |
|---|---|---|---|---|
| Levenshtein | 1.00 | 0.93 | 0.96 | 0.93 |
| Monge Elkan | 0.94 | 0.96 | 0.95 | 0.89 |
| TFIDF | 0.98 | 0.91 | 0.94 | 0.89 |
Levenshtein and Monge-Elkan were given higher weights because most of the schema elements varied in their spelling and token order like ‘salt 1 units’ and ‘salt unit1’. The TF*IDF weight helped find matches like ‘salt’ with ‘salt_name’ rather than ‘salt_conc’ or ‘salt_unit’ because the tokens ‘conc’ and ‘unit’ appear more frequently in the input schema than the ‘name’ token. Hence, the ‘name’ token has higher TF*IDF weight.
Algorithm for 1:1 Matching:
A similarity matrix for headers is generated with values in the range [0, 1] where 1 indicates a perfect linguistic match. After calculating the similarity matrix, the best match for each element was found based on the maximum similarity value of an element. The match generation algorithm is provided in Algorithm 1.
| 10–11: | A threshold of 0.5 is applied to the similarity values for each input header to find the candidate output headers and the candidates are sorted by their descending similarity values. |
| 12–14: | For each input header, our algorithm finds the output header with the highest similarity value. |
| 15–17: | If the output header is not used for any previous match, the match between the current input header and output header is taken while storing its similarity value. |
| 18–20: | If the output header has already been used for a previous input header (prevMatch in the algorithm), their similarity values are compared. |
| 21–24: | If the current input header has a higher similarity value, its match is accepted while discarding the match of previous input header. Then a match for this previous input header is found in a recursive way. |
| 12–14: | If the current input header has a lower or equal similarity value, it is matched with the next candidate output header following the same rules (continuation of the for loop in the algorithm). |
Algorithm 1.
Match Generation Algorithm
| 1: | Input: input_headers, output_headers, simMatrix |
| 2: | Output: matches |
| 3: | matches ← empty list |
| 4: | for all input ∈ input headers do |
| 5: | MATCHINPUT(input, matches) |
| 6: | end for |
| 7: | return matches |
| 8: | |
| 9: | procedure MATCHINPUT(input, matches) |
| 10: | ops ← output headers with similarity values >= 0.5 |
| 11: | sort ops by descending similarity value |
| 12: | for all output ∈ ops do |
| 13: | sim ← simMatrix[input][output] |
| 14: | currMatch ← [input, output, sim] |
| 15: | if output is not already matched then |
| 16: | matches.add(currMatch) |
| 17: | return |
| 18: | else |
| 19: | find prevMatch of output from matches |
| 20: | if prevMatch.sim < currMatch.sim then |
| 21: | matches.add(currMatch) |
| 22: | matches.remove(prevMatch) |
| 23: | MATCHINPUT(prevMatch.input, matches) |
| 24: | return |
| 25: | end if |
| 26: | end if |
| 27: | end for |
| 28: | return |
| 29: | end procedure |
Our method enforces 1:1 mapping. If an input header has the same maximum similarity value with more than one output header, the first output header among them is chosen. This works as we have designed the intermediate schema such that the prioritized headers appear first in the list. This match generation procedure results in a 1:1 match for the headers in the input schema with those in the intermediate schema. Due to the use of a threshold to ignore the similarity values less than 0.5, the relationship between the input and the intermediate schema is not total (i.e., some of the headers in a schema may not have a matching element in the other schema).
Refining matches:
This step includes resolving ambiguity in chemical group matching and ambiguity in other matches, using a dictionary for domain specific matches, and obtaining user feedback.
Ambiguity in Chemical Group Matching.
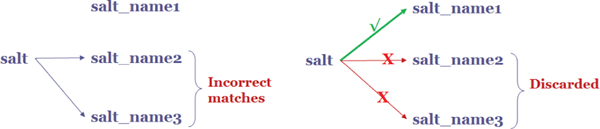
If the input schema contains only one salt or one precipitant then there may not be a digit suffix in their names, for example, ‘salt’. This ‘salt’ can be matched with any of ‘salt_name1’, ‘salt_name2’ or ‘salt_name3’ in our intermediate schema. Algorithm 1 prioritizes the matching of ‘salt’ with ‘salt_name1’, due to its order. If there are cases where the similarity value with ‘salt_name2’ or ‘salt_name3’ may be higher due to the TF*IDF weight in similarity metrics (the numbers 2 or 3 may have higher weight due to their low frequency in the input schema). In such cases, the matches are refined to match columns with a lower numbered suffix than a higher numbered suffix.
Resolving Ambiguous Matches.
In the mappings list, if none of ‘salt_name1’, ‘salt_conc1’ and ‘salt_unit1’ is matched but any of ‘salt_name2’/‘salt_name3’, ‘salt_conc2’/’salt_conc3’ or ‘salt_unit2’/’salt_unit3’ is matched, the matched header from the input schema is taken and updated to match ‘salt_name1’ for name, ‘salt_conc1’ for conc and ‘salt_unit1’ for unit columns as shown in Fig 6. If there are matches for salt2 as well as salt3 but not for salt1, the matches for salt2 are moved to match with salt1 and salt3 matches moved to salt2. The same refining process is performed for precipitants as well. Hence, the resulting mapping list contains the matches prioritized by the digit suffix.
Fig. 6.
Correcting ambiguous matches
Building and mapping screen domain representations.
A domain dictionary is used to bring uniformity in the words by expanding abbreviations and mapping words to their domain-specific meanings, for example, ‘ppt’ to ‘precipitant’, ‘additive’ to ‘salt’, ‘tube’/’screen’ to ‘well’, ‘#’/’no’ to ‘id’. When the available input screens were analyzed further, we found that ‘[Salt]’ referred to ‘salt conc’ and ‘[Salt] units’ referred to ‘salt units’. These mappings are also handled for tokens of each header.
User Feedback.
Further refinement is done by taking user feedback for the generated matches. The matches generated can be accepted or rejected by the user or even new matches can be added for the particular screen file(s). The accepted matches are saved by the system for reuse.
C. Data Integration
The second phase of the research transforms the data in the given input screen to the output screen format. We use the output of the first phase, i.e., list of mappings between the schemas for data transformation. The output screen can be in a predefined format used by the analysis and visualization programs or a customized format uploaded by the user. Our system is able to analyze headers of screens provided by the user (i.e., user defined) and perform mapping based on these headers. For the predefined screen files, the mappings between the intermediate and output schema can be preset ahead of time, and the system only finds the mappings between input and intermediate schema using the linguistic matching process. For the customized output screens, input-to-intermediate and output-to-intermediate mappings should be generated [9].
Transforming Cocktails from Input File to Intermediate File for Matching Headers:
If a match for a header is not found in the mapping list, NULL is added instead of its index to the list. Then the rows of the input file are iterated over, copying data from input to intermediate file by rearranging the cells based on the list of indices. For example, the first few headers in the intermediate schema are ‘well_id’, ‘buffer_name’, ‘buffer_conc’, ‘buffer_unit’, etc. If we match Anatrace MCSG screen (Fig. 1) with the intermediate schema, the list of indices will be [1, 7, 5, 6,...] using 0-based index (starting at 0). So the data is copied from column 1 of the input file to the first column in intermediate file, column 7 of the input file to the second column in the intermediate, column 5 to the third, column 6 to the fourth, and so on.
Splitting Composite Chemical Names and Properties in Input File:
Input screens like Qiagen (Fig. 3) do not have columns separated as ‘conc’, ‘unit’, ‘name’ or ‘ph’. They only have one column header for each group of chemicals (Salt, Buffer and Precipitant), and the values in these columns contain a combination of conc, unit, name and ph values. Such columns are parsed to find the appropriate conc, unit, name and ph values and placed under their own headers in the intermediate file. In our intermediate schema headers, there are 6 groups of chemicals: buffer, salt1, salt2, salt3, precipitant1 and precipitant2. Their indices groups in the intermediate schema are 1–4, 5–7, 8–10, 11–13, 14–16 and 17–19 respectively. For each group of indices, it is checked whether only one out of three (out of four for buffer) indices in a group was a non-NULL value and analyzed if parsing data is needed. The screen files contain the grouped data in the format “<conc> <unit> <chemical name>” for salts and precipitants and “<conc> <unit> <chemical name> ph <ph value>” for buffers. So, the contents of the data columns are split by spaces and the values are assigned to their respective columns in the intermediate file. For example, if the data column of buffer type has value “0.1 M Citric acid pH 5.0”, then the contents are separated as ‘buffer_conc’ = ‘0.1’, ‘buffer_unit’ = ‘M’, ‘buffer_name’ = ‘Citric acid’ and ‘buffer_ph’ = ‘5.0’.
Headers in the Predefined Output Format:
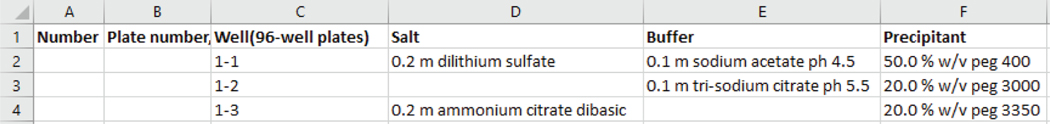
The predefined output format contains a fixed set of headers designed in the lab for use in analysis programs like AED [1], [3], GenScreen [4], [5], and Visual-X2 [6]. In our representation, we opt to split a chemical into its anion and cation where possible. The headers present in this format are ‘Well_Id’, ‘B_Anion’ (or B), ‘B_Cation’, ‘Ph’, ‘B_Conc’, ‘C1_Anion’ (or C1), ‘C1_Cation’, ‘C1_Conc’, ‘C1_M’, ‘C1_Ph’, ‘C2_Anion’ (or C2), ‘C2_Cation’, ‘C2_Conc’, ‘C2_M’, ‘C2_Ph’, ‘C3_Anion’ (or C3), ‘C3_Cation’, ‘C3_Conc’, ‘C3_M’, ‘C3_Ph’, ‘C4_Anion’(or C4), ‘C4_Cation’, ‘C4_Conc’, ‘C4_M’, ‘C4_Ph’, ‘C5_Anion’ (or C5), ‘C5_Cation’, ‘C5_Conc’, ‘C5_M’, ‘C5_Ph’, ‘S_a’, ‘S_b’, ‘S_c’. This representation is considered handy for the biochemist and for analyzing data. ‘Well_id’ is an identifier of the cocktail and also refers to the position of the condition in the experiment plate. The next headers can be divided into a reagent type and its property separated by an underscore. There are 6 reagent groups: B, C1, C2, C3, C4, C5. The column B refers to the buffer reagent, C1 is the precipitant and C2, C3, C4 and C5 are additives which can be salts or precipitants. ‘M’ and ‘Conc’ refers to the concentration of the chemicals in Molarity and other units respectively. The ‘Ph’ column contains the ph value of the reagent. The reagents in the intermediate headers map to the output header columns as buffer –> B, precipitant1 –> C1, salt1 –> C2, precipitant2 –> C3, salt2 –> C4, and salt3 –> C5. The last three columns ‘S_a’, ‘S_b’, ‘S_c’ refers to the ranking of the cocktail generated by the result of the experiments in the wet lab and hence is left empty by our program.
Splitting Salts into Anions and Cations and Assigning Unit Values for Predefined Output File:
Based on these predefined mappings, the intermediate file is converted to the output format following the similar process as in the input to intermediate file conversion. In the predefined output format, a name column is split into anion and cation columns. Chemicals such as polymers and detergents cannot be clearly separated into cations and anions. In such cases, the chemical name resides in the ‘_Anion’( or ‘Chemical’) column, with the’_Cation’ column remaining empty. The output file does not contain a separate column for unit values. For a reagent whose unit value is ‘M’ (molar), the conc value is placed under the ‘M’ type column and for other units, the conc value placed under the ‘conc’ type column.
Mapping to Customized Output File and Generating Composite Chemical Names and Properties:
A customized output is a format uploaded by the user. The header names are not fixed so predefined mappings cannot be used for data transformation. In this case, the output-to-intermediate mappings generated by our match generation algorithm in Algorithm 1 are used. First, a list of indices or column positions is formed by finding a matching intermediate header for each output header. Then the rows of the intermediate file are iterated over copying each row by rearranging the columns based on the list of indices. However, this method is not enough for output screen files like Qiagen, which do not have conc, units and name values separated across columns. Since the output file only contains one header column for each reagent, the data from conc, name, unit and ph columns should be merged together while writing to the output file. For this, the header groups which required merging are determined following similar methods as above in parsing. Then the data columns of intermediate headers are merged to the available column in the output schema in the format “<conc> <unit> <chemical name>” for salts and precipitants and “<conc> <unit> <chemical name> ph <ph value>” for buffers.
D. Consistent Naming of Chemicals
The output screen file should have consistent names of chemicals. Different companies may use different identifiers as chemical names and there are multiple names used to represent a chemical. Some of the chemical identifier representations are MOL, SMILES notation, InChI strings, IUPAC names, etc. These representations are called systematic identifiers, which are generated algorithmically and should have a one-to-one correspondence with the structure (however different software could generate different formats) [27].
Consistent naming is a challenging task. The issue is not only the use of a standard naming convention. Some of these naming conventions look weird (and produce lengthy descriptions) and are different from daily use of these chemical names. To overcome this problem, we allowed the expert to use his or her naming for a chemical while using standard naming conventions for mapping chemicals among screens. The display_name is the chemical name favored by the expert for the output file in our analysis.
In order to systematize the naming of all the compounds by avoiding assignment of two compounds to the same name, IUPAC naming rules are followed. IUPAC names are based on the structure of the compound. We used IUPAC names among others to map a chemical to a consistent name in our research as IUPAC names are also more readable and understandable by humans. For each chemical name in the input screen file, its IUPAC name is found by looking it up in Chemical Identifier Resolver (CIR) website5. CIR works as a resolver for different chemical structure identifiers and allows one to convert a given structure identifier into another representation or structure identifier. We used a python wrapper named cirpy [28] which handles constructing URL requests and parsing XML responses from the CIR website. If a chemical name could not be resolved by this resolver, the chemical name itself is used as its identifier.
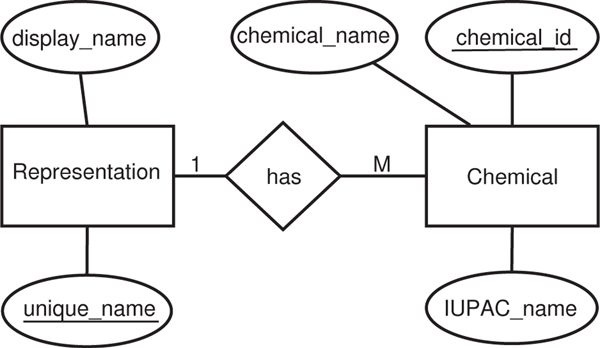
We created a database with two tables named ‘chemical’ and ‘representation’ as shown in the entity-relationship diagram in Fig. 7. The ‘chemical’ table contains 4 fields: ‘chemical_id’ (the auto-incremented key), ‘chemical_name’, ‘IUPAC_name’ and ‘unique_name’. The ‘representation’ table contains two fields: ‘unique_name’ (the primary key for this table and foreign key in the ‘chemical’ table) and ‘display_name’ (set to NULL by default). The display_name is equivalent to the chosen name by the user. In the ‘chemical’ table, we have all the chemical names as they appear in the screen files and their corresponding IUPAC names. Each chemical name is first mapped to its IUPAC name and saved to the ‘chemical’ table and the IUPAC name is added to the ‘representation’ table, if not present. Then, if a new IUPAC name comes up, for which user has not chosen a display_name, the user is given an option to choose a display_name for it. The user is provided a list of all possible names seen so far in the screen files for that IUPAC name and allowed to choose one of them as its display_name or even enter a new name outside of the list. This chosen display name is saved in the ‘representation’ table for reuse automatically when the chemical name is encountered.
Fig. 7.
ER diagram for chemical names representation
During the transformation of the intermediate file to the output screen file, the display_name of each chemical is found from the database tables ‘chemical’ and ‘representation’ and the chemical name is replaced with its display name in the output screen file. Since multiple representations of a chemical map to the same IUPAC name, they will have a single display name. Hence, the output screen file contains the standard names of the chemicals as defined by the experts.
After applying naming consistency to the screen, the output screen is available for download through the user interface. An example of output screen file of JCSG screen is shown in predefined format Fig. 8 and in Qiagen format in Fig. 9.
Fig. 8.
Snapshot of output file for JCSG screen in predefined format
Fig. 9.
Snapshot of output file for JCSG screen in Qiagen format
IV. Experiments and Results
In order to evaluate the results of schema matching and data integration, we carried out a number of experiments with different screen files. We first solved the matching task manually to compare the results with the automatic approach. We implemented our program as a web application which takes an input screen file (and an output screen file for user-defined output) in MS Excel format and outputs an MS Excel file in specified format. After the mappings are proposed by the system, the user has the ability to accept or discard the match and even add new matches. We evaluate the quality of our system in two stages separately for the schema matching phase and data integration phase.
A. Crystal Screens Data
We have collected data from many commercial screen providers including Anatrace, Axygen, Fluidigm, Hampton Research, Jena BioScience, KeraFAST, Molecular Dimensions, Omscientia, QiaGen, Rigaku Reagents, Sigma, and XtalQuest. We have tested > 300 crystal screens files, which were mostly publicly accessible though the vendors’ websites. If screen files are available in PDF format, the files should be converted into MS Excel format.
B. Results
1). The ScreenMatcher Web Tool:
The method described above has been implemented as a stand alone web service available at http://datamedia.cs.uah.edu/screenmatcher/main. The primary code was written in Python 2.7 and Microsoft SQL server has been used for managing the database. The ScreenMatcher web tool allows the user to upload a source screen file, in MS Excel format, then it automatically matches the source header with the target header, and completely transfers data into the target file format. The ScreenMatcher has a default output format, however it allows users to use their custom format as well. The ScreenMatcher has two interfaces: schema matching and consistent naming.
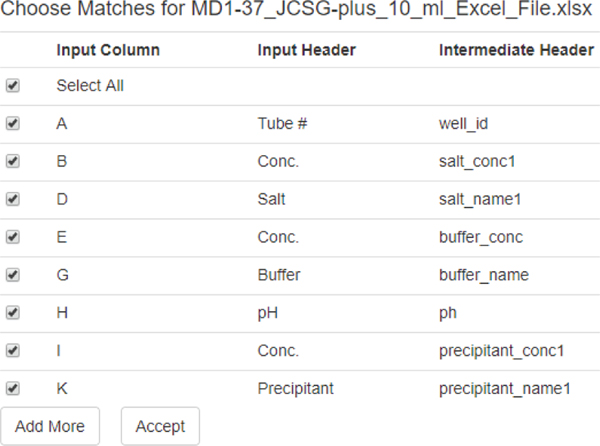
Figure 10 shows a snapshot of header matching review for JCSG screen from Molecular Dimensions. The system provides found matches between headers with the option of selecting or ignoring matched headers while adding more or different matches by the user.
Fig. 10.
The ScreenMatcher Web tool: Matching snapshot
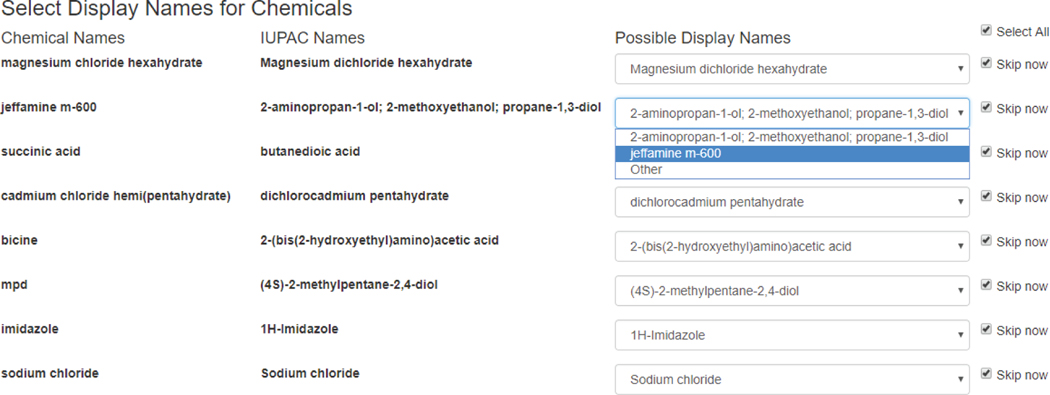
If a chemical name appears first time in the analysis, the system provides the IUPAC name (if available) along with the original chemical name. For each chemical, it provides possible display names or the user may provide a custom name to be used in the research laboratory. Figure 11 provides a snapshot of selecting display names for chemicals for JCSG screen from Molecular Dimensions. By selecting skip now check box, the user may skip selecting display names at the moment and review again next time when they appear. To evaluate the naming consistency we manually compared the chemical names between input and output files to ensure that the chemical names were properly converted.
Fig. 11.
The ScreenMatcher Web tool: consistent naming
2). Evaluation of Schema Matching:
We have evaluated > 300 crystal screens. Since the screens from the same company share similar headers, our system detects the header format and use the same mappings if a screen file format is matched previously. Using more number of screens do not contribute to the schema matching.
For quantitative analysis, we have used 10 screens which have different header formats. The screens are AmSO4 Suite Composition and The Classics II Suite Composition from Qiagen6, Anatrace - Microlytic MCSG1 Formulations from Anatrace7, CS-101L JBScreen Classic 1 from Jena Bio-Science8, HR007407 Hampton Crystal Screen Formulation from Hampton research9, and MD1–13 3D Structure Screen, MD1–02 Structure Screen, MD1–37 JCSG-plus, MD1–104 The BCS Screen, and MD1–35 from Molecular Dimensions10. Table II presents the correctness of header match in 10 screens. The headers in the table are Screen name, TP (True Positive), FP (False Positive), FN (False Negative), P (Precision), R (Recall), F (F1-measure) and O (Overall). We can observe that the number of correct matches is high for all screens and the number of incorrect matches is 0. Two of the screens missed 1 intended match. The Classics screen has 4 salts and 1 precipitant but since our intermediate schema accommodates only 3 salts, one of them was missed by the system. This error can be resolved by accommodating more salts in the schema. In JBScreen, the match for the column ‘well_id’ was missed because these screens do not use identifiers present in our domain dictionary. Since the ‘well_id’ field has less importance than other reagent fields, we can say that our system works well with almost all the screens tested.
TABLE II.
Experimental Results of 10 screens
| Screen | TP | FP | FN | P | R | F | O |
|---|---|---|---|---|---|---|---|
| AmSO4 | 5 | 0 | 0 | 1 | 1 | 1 | 1 |
| Classics II | 6 | 0 | 1 | 1 | 0.86 | 0.92 | 0.86 |
| MCSG1 | 14 | 0 | 0 | 1 | 1 | 1 | 1 |
| CS-101L | 12 | 0 | 1 | 1 | 0.92 | 0.96 | 0.92 |
| HR007407 | 14 | 0 | 0 | 1 | 1 | 1 | 1 |
| MD1–13 | 10 | 0 | 0 | 1 | 1 | 1 | 1 |
| MD1–02 | 20 | 0 | 0 | 1 | 1 | 1 | 1 |
| MD1–37 | 8 | 0 | 0 | 1 | 1 | 1 | 1 |
| MD1–104 | 17 | 0 | 0 | 1 | 1 | 1 | 1 |
| MD1–35 | 20 | 0 | 0 | 1 | 1 | 1 | 1 |
3). Data Integration:
For the data integration phase, we use the completeness measure to assess the quality of the results. Completeness is a concept from data integration and migration which attempts to identify objects that are missing in the target file that were present in the source file [29]. To evaluate the data integration phase, the input and output files were compared manually to ensure that all the rows from input file were transferred to the output file and also all header column present in the mappings were present in the output file. In our observations, our system did not to mislabel any chemical (in our analysis) and all rows were properly mapped.
C. Comparison with other matchers
We also compared our matcher with two other matchers: the Harmony matcher [19] and the FlexMatcher [20]. We tested the same six commercial screens that we used for comparing similarity measures to compare our system with Harmony and FlexMatcher.
Harmony matcher has a GUI which allows the users to import the schema in different formats and generates mappings for the schemas. The mappings are shown as color-coded lines connecting the source and target elements with the colors representing their confidence scores. We imported our screen files as spreadsheet documents to the Harmony matcher and ran the name and documentation matchers of Harmony. The mappings generated by the system were m:n, i.e., an element from the source schema was matched with multiple elements from the target schema with the same score and vice-versa. Although the matcher recognized spreadsheet documents, it ignored the multiple columns with the same names and treated them as one. Fig. 12 shows the Harmony matcher’s result of mappings between the Classics screen and our intermediate schema. We can see that there are multiple matches for an element in the input schema but we considered those as correct matches if any of them was correct.
Fig. 12.
Snapshot of Harmony matcher when mapping the Classics screen and our intermediate schema.
We developed the code to create dataframes with the screens data and trained FlexMatcher with 4 screens and tested them individually with 6 screens. In the training phase, we created the dataframes from the header row and four data rows for each screen. Then we defined the mappings from each of the four schemas to the intermediate schema. Then the classifier was trained with the dataframes and the mappings. For testing, we created the dataframes using header and data rows for each of the six screens and predicted the mappings for them. The output of the program was a dictionary of mappings from input to intermediate schema. The mappings were n:1, i.e., multiple elements in the source schema could be matched with the same target schema element. This matcher also did not accept multiple columns with the same name so we modified some of the headers by adding digit suffixes to make them unique.
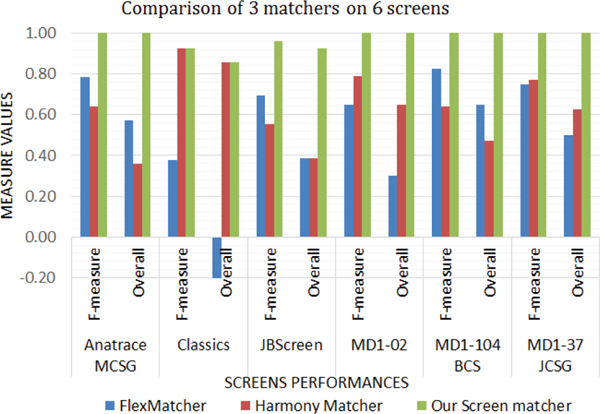
We recorded the results of the matchers for each of the screens and tested our screen matcher with the same screens. The results of F1-measure and overall for each screen with 3 matchers are shown in Fig. 13. We label our matcher as ‘screen matcher’ in the tables and figures. We can see that our screen matcher has the best accuracy for all the screens.
Fig. 13.
Comparison of our screen matcher with FlexMatcher and Harmony across 6 screens
Table III shows the combined results of the values of the evaluation measures for the three matchers (FlexMatcher, Harmony and Screen Matcher) across six commercial screens. The FlexMatcher had an overall accuracy of 40%, Harmony had an overall accuracy of 53% while our screen matcher had an overall accuracy of 97%. Hence our screen matcher is the most applicable in the field of protein crystallization screens.
TABLE III.
Comparison of evaluation measures for three matchers
| Matcher names | Precision | Recall | F-measure | Overall |
|---|---|---|---|---|
| FlexMatcher | 0.70 | 0.70 | 0.70 | 0.40 |
| Harmony Matcher | 0.94 | 0.57 | 0.71 | 0.53 |
| Screen Matcher | 1.00 | 0.97 | 0.99 | 0.97 |
D. Discussion
Our mapping between screen files is based on an intermediate schema. Our intermediate schema had 6 groups of chemicals: one buffer, three salts, and two precipitants. This schema had a sufficient number of groups for many screens in our analysis. The number of chemical groups may be increased if input screens have more types of chemicals.
Headers may appear in any row in a spreadsheet. We identify data rows to determine where headers are. Our assumption is that the headers should be just before the data row. In some cases, the headers occupied two rows as shown in Fig. 1. For multi-row headers, we observed that the first row had the actual header and the second row for the header had extra information. We have checked occupancy of the rows to determine whether headers occupy two rows or not. We have not observed any screen file having headers occupying more than two rows. We assume that the headers only appears once in the file and before the data rows.
For repetitive attribute problem, we firstly determined the types of attributes and identified as property attributes such as concentration. These attributes are typically located close to their chemical; however, they could appear before or after the chemical name. If the properties of the first chemical appear early (before the chemical name), then those repetitive attributes precede the chemical name or vice versa.
Our screen matcher tool provides matches with high accuracy. It also provides full control of matching by accepting, rejecting, or adding matches. Even if screen files do not have proper headers for columns or they have empty columns, the user may match empty column headers to our intermediate headers and the system can generate output files in the desired output format. Our schema matcher tool also enables mapping between any two screen file formats. This matching is done once, and our tool learns the matches afterwards.
We have also tried to bring consistency in the naming of the chemicals across different screen files by mapping each chemical to their IUPAC names using the CIR website. However, not all chemicals were recognized by this resolver. Our method of resolving chemical names can be improved by using multiple resolvers so that chemical names which are not recognized by one can be resolved by the others. If the commercial companies provide ambiguous chemical names, our tool does not resolve those ambiguities. We rather provide a consistent naming framework for the expert so that the expert can choose preferred names or categories and regenerate screen files with consistent names. For the long run, data standard initiatives with broad industry participation, as for example in mass-spectrometry for proteomics, are the best way for confronting these difficulties.
V. Conclusion
This paper presents an approach to solve the problem of discrepancies in the protein screens of different companies. We used the linguistic schema matching method which uses the element names of the schema to find the matches based on their similarity values. We also mapped the chemical names to their consistent names chosen by the user and used those names in the output files. Our matcher accepts input in MS Excel format so there is no need to define the schema in schema definition languages. It generates 1:1 mappings between the source and target schema handling the cases of matches having same similarity scores based on priority. Furthermore, it also handles the ambiguous names in the schema by looking at the structure of the file and grouping the names of related elements. We also compared our approach with two matchers (Harmony and FlexMatcher) using six screen files and observed that our screen matcher had 97% accuracy compared to 40% accuracy of FlexMatcher and 53% accuracy of Harmony matcher. We have applied our matcher to the protein crystallization screens. We believe this algorithm can also be applicable to other domains with minor modifications.
Acknowledgment
This research was supported by National Institutes of Health (GM116283) grant.
Biography

Midusha Shrestha received her Bachelor’s degree in Computer Engineering from Institute of Engineering, Central Campus, Nepal in 2014, and an M.S. degree in Computer Science from the University of Alabama in Huntsville in 2017. Her M.S. thesis is related to schema matching and transformation of commercial screens used in protein crystallization. Her research interests include machine learning, data mining and analytics, and database systems.

Truong X. Tran received his Bachelor’s degree major in Electronic and Telecommunication Engineering, minor in Biomedical Electronic Engineering, from Hanoi University of Science and Technology, Vietnam in 2010, the M.S. degree in Computer Science from Arkansas State University, AR, USA in 2012. He is pursuing Doctoral degree in Computer Science at the University of Alabama in Huntsville, AL, USA. He works in Data analysis for protein crystallization research project. His research interests include machine learning and data mining.

Bidhan Bhattarai received his Bachelor’s degree in Computer Engineering from Institute of Engineering, Central Campus, Nepal in 2015, and an M.S. degree in Computer Science from the University of Alabama in Huntsville in 2017. His M.S. thesis is related to applying genetic algorithm to determine factors crucial for successful protein crystallization. His research interests include deep learning, neural networks, and natural language processing.

Marc L. Pusey is a research scientist working at iXpressGenes, Inc., Huntsville Alabama. Dr. Pusey received his Ph.D. in Biochemistry from the University of Miami, Fl., then did post-doctoral research at the University of Minnesota. He moved to Huntsville in 1985, and worked at NASA/MSFC for the next 23 years in the new field of protein crystal growth. After retirement from NASA, he obtained his current position at iXpressGenes with the focus of his research being improved methods for protein crystal screening, crystal detection, and crystallization condition identification using visible fluorescence methods.

Ramazan S Aygün: received the B.S. degree in computer engineering from Bilkent University, Ankara, Turkey in 1996, the M.S. degree from Middle East Technical University, Ankara in 1998, and the Ph.D. degree in computer science and engineering from State University of New York at Buffalo in 2003. He is currently an Associate Professor in Computer Science Department, University of Alabama in Huntsville. His research interests include protein crystallization image analysis, data mining, image and video processing, spatio-temporal indexing and querying, multimedia databases, semantic computing, multimedia networking, and multimedia synchronization.
Footnotes
Contributor Information
Midusha Shresta, Data Media Lab, Computer Science Department, University of Alabama in Huntsville, Huntsville, Alabama 35899, United States.
Truong X. Tran, Data Media Lab, Computer Science Department, University of Alabama in Huntsville, Huntsville, Alabama 35899, United States
Bidhan Bhattarai, Data Media Lab, Computer Science Department, University of Alabama in Huntsville, Huntsville, Alabama 35899, United States.
Marc L. Pusey, iXpressGenes, Inc., Huntsville, Alabama 35806, United States
Ramazan S. Aygun, Data Media Lab, Computer Science Department, University of Alabama in Huntsville, Huntsville, Alabama 35899, United States.
References
- [1].Dinc I, Pusey ML, and Aygun RS, “Optimizing associative experimental design for protein crystallization screening,” IEEE Transactions on NanoBioscience, no. 99, p. 1, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sigdel M, Pusey ML, and Aygun RS, “Real-time protein crystallization image acquisition and classification system,” Crystal growth & design, vol. 13, no. 7, pp. 2728–2736, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dinc I, Pusey ML, and Aygün RS, Protein Crystallization Screening Using Associative Experimental Design. Cham: Springer International Publishing, 2015, pp. 84–95. [Online]. Available: 10.1007/978-3-319-19048-8_8 [DOI] [Google Scholar]
- [4].Acharya S, “GenScreen : A genetic algorithm for protein crystallization screening,” Master’s thesis, The University of Alabama in Huntsville, Huntsville, Alabama, USA, 2017. [Google Scholar]
- [5].Bhattarai B, Shrestha M, Aygun RS, and Pusey ML, “Optimizing genetic algorithm for protein crystallization screening using an exploratory fitness function,” in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), November 2017, pp. 2083–2090. [Google Scholar]
- [6].Subedi S, Pusey ML, and Aygun RS, “Visual-x2: Scoring and visualization tool for analysis of protein crystallization trial images,” in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), November 2017, pp. 2316–2318. [Google Scholar]
- [7].Hedderich T, Marcia M, Kpke J, and Michel H, “Pickscreens, a new database for the comparison of crystallization screens for biological macromolecules,” Crystal Growth & Design, vol. 11, no. 2, pp. 488–491, 2011. [Online]. Available: 10.1021/cg101267n [DOI] [Google Scholar]
- [8].Newman J, Fazio VJ, Lawson B, and Peat TS, “The c6 web tool: A resource for the rational selection of crystallization conditions,” Crystal Growth & Design, vol. 10, no. 6, pp. 2785–2792, 2010. [Online]. Available: 10.1021/cg1004209 [DOI] [Google Scholar]
- [9].Shrestha M, Bhattarai B, Aygun RS, and Pusey ML, “Schema matching and data integration on protein crystallization screens,” in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), November 2017, pp. 2306–2308. [Google Scholar]
- [10].“Data Integration Info: Quick view on world of data,” http://www.dataintegration.info/, accessed: 2017-07-28.
- [11].Newman J, Peat TS, and Savage G, “What’s in a Name? Moving Towards a Limited Vocabulary for Macromolecular Crystallisation,” Australian Journal of Chemistry, vol. 67, no. 12, p. 1813, Jul. 2014. [Google Scholar]
- [12].Lenzerini M, “Data integration: A theoretical perspective,” in Proceedings of the Twenty-first ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems, ser. PODS ‘02. New York, NY, USA: ACM, 2002, pp. 233–246. [Online]. Available: http://doi.acm.org/10.1145/543613.543644 [Google Scholar]
- [13].Gorrec F, “Protein crystallization screens developed at the MRC laboratory of molecular biology,” Drug Discovery Today, vol. 21, no. 5, pp. 819–825, 2016. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S1359644616300794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Unal O, Afsarmanesh H et al. , “Using linguistic techniques for schema matching.” in ICSOFT (2), 2006, pp. 115–120. [Google Scholar]
- [15].Madhavan J, Bernstein PA, and Rahm E, “Generic Schema Matching with Cupid,” in Proceedings of the 27th International Conference on Very Large Data Bases, ser. VLDB ‘01. San Francisco, CA, USA: Morgan Kaufmann Publishers Inc., 2001, pp. 49–58. [Online]. Available: http://dl.acm.org/citation.cfm?id=645927.672191 [Google Scholar]
- [16].Melnik S, Garcia-Molina H, and Rahm E, “Similarity flooding: A versatile graph matching algorithm and its application to schema matching,” in 18th International Conference on Data Engineering (ICDE 2002), 2002. [Online]. Available: http://ilpubs.stanford.edu:8090/730/ [Google Scholar]
- [17].Do HH and Rahm E, “COMA: A system for flexible combination of schema matching approaches,” in Proc. 28th Int. Conf. on Very Large Data Bases VLDB Endowment, 2002, pp. 610–621. [Google Scholar]
- [18].Do HH, “Schema Matching and Mapping-based Data Integration,” Ph.D. dissertation, University of Leipzig, Germany, August 2005. [Online]. Available: http://lips.informatik.uni-leipzig.de/files/2006-4.pdf [Google Scholar]
- [19].Mork P, Rosenthal A, Korb J, and Samuel K, “Integration workbench: Integrating schema integration tools,” in 22nd International Conference on Data Engineering Workshops (ICDEW’06), 2006, pp. 3–3. [Google Scholar]
- [20].“Schema Matching and Merging — BigGorilla,” https://www.biggorilla.org/schema-matching-and-merging, accessed: 2017-08-21.
- [21].Neville J and Jensen D, “Supporting relational knowledge discovery: Lessons in architecture and algorithm design,” in Proceedings of the Data Mining Lessons Learned Workshop, 19th International Conference on Machine Learning, 2002. [Google Scholar]
- [22].Levenshtein VI, “Binary codes capable of correcting deletions, insertions, and reversals,” in Soviet physics doklady, vol. 10, no. 8, 1966, pp. 707–710. [Google Scholar]
- [23].Monge AE, Elkan C et al. , “The Field Matching Problem: Algorithms and Applications.” in KDD, 1996, pp. 267–270. [Google Scholar]
- [24].Rijsbergen CJV, Information Retrieval, 2nd ed. Newton, MA, USA: Butterworth-Heinemann, 1979. [Google Scholar]
- [25].Doan A, Halevy A, and Ives Z, Principles of Data Integration. 225 Wyman Street, Waltham, MA 02451, USA: Morgan Kaufmann, 2012, ch. 4. [Google Scholar]
- [26].Jaccard P, “The distribution of the flora in the alpine zone.1,” New Phytologist, vol. 11, no. 2, pp. 37–50, 1912. [Online]. Available: 10.1111/j.1469-8137.1912.tb05611.x [DOI] [Google Scholar]
- [27].Akhondi SA, Kors JA, and Muresan S, “Consistency of systematic chemical identifiers within and between small-molecule databases,” Journal of Cheminformatics, vol. 4, no. 1, p. 35, December 2012. [Online]. Available: 10.1186/1758-2946-4-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].“CIRpy - A Python interface for the Chemical Identifier Resolver (CIR),” http://blog.matt-swain.com/post/19633070138/cirpy-a-python-interface-for-the-chemical, accessed: 2017-09-01.
- [29].Matthes F, Schulz C, and Haller K, “Testing & quality assurance in data migration projects,” in 2011 27th IEEE International Conference on Software Maintenance (ICSM), September 2011, pp. 438–447. [Google Scholar]