Abstract
Type 1 diabetes (T1D) affects over 200,000 children and is associated with an increased risk of cognitive dysfunction. Prior imaging studies suggest the neurological changes underlying this risk are multifactorial, including macrostructural, microstructural, and inflammatory changes. However, these studies have yet to be integrated, limiting investigation into how these phenomena interact. To better understand these complex mechanisms of brain injury, a well-powered, prospective, multisite, and multimodal neuroimaging study is needed. We take the first step in accomplishing this with a preliminary characterization of multisite, multimodal MRI quality, motion, and variability in pediatric T1D. We acquire structural T1 weighted (T1w) MRI, diffusion tensor MRI (DTI), functional MRI (fMRI), and magnetic resonance spectroscopy (MRS) of 5-7 participants from each of two sites. First, we assess the contrast-to-noise ratio of the T1w MRI and find no differences between sites. Second, we characterize intervolume motion in DTI and fMRI and find it to be on the subvoxel level. Third, we investigate variability in regional gray matter volumes and local gyrification indices, bundle-wise DTI microstructural measures, and N-acetylaspartate to creatine ratios. We find the T1-based measures to be comparable between sites before harmonization and the DTI and MRS-based measures to be comparable after. We find a 5-15% coefficient of variation for most measures, suggesting ~150-200 participants per group on average are needed to detect a 5% difference across these modalities at 0.9 power. We conclude that multisite, multimodal neuroimaging of pediatric T1D is feasible with low motion artifact after harmonization of DTI and MRS.
Keywords: type 1 diabetes, multimodal MRI, variability, motion, cortical morphometry, diffusion microstructure, fiber tractography, magnetic resonance spectroscopy
1. INTRODUCTION
Type 1 diabetes (T1D) affects almost 1.7 million Americans, including over 200,000 children and adolescents, and is associated with an increased risk of mild to moderate cognitive dysfunction [1], [2]. Currently, the neurological changes that underlie this risk are thought to be multifactorial. Prior studies have pointed to neuroanatomical macrostructural [3], [4], microstructural [5], and inflammatory changes [6], [7], but these studies have yet to be integrated into a consistent framework, limiting investigation into how these phenomena interact. To better understand these complex mechanisms of brain injury, a well-powered, prospective, multisite, and multimodal magnetic resonance imaging (MRI) study is needed.
The success of such a study depends on acquiring high quality images with low motion artifact in a pediatric cohort from multiple sites and scanners. This requires pediatric participants to remain still in the MRI scanner for the duration of imaging and the consideration of interscanner variability that could confound such an analysis [8]. As such, in this work, we present a pilot study to assess multisite and multimodal MRI quality, motion, and variability; characterize how two common harmonization techniques can reduce some interscanner biases in pediatric T1D; and investigate the feasibility of a large-scale multisite, multimodal neuroimaging study for pediatric T1D.
We first acquire multimodal neuroimaging, including structural, diffusion, functional, and spectroscopic MRI in a small cohort of pediatric T1D participants at two sites, each with a different MRI scanner. We explore image contrast, motion artifacts, and how quantitative metrics extracted from the images vary within and between the two sites. Last, using these results, we perform a sample size analysis to determine the number of T1D participants and healthy controls needed to resolve a 5% difference between groups across these modalities at scale.
2. METHODS
2.1. Study design and MRI acquisitions
Our study population consists of 12 pediatric participants with T1D imaged across two sites: site A (N = 7, 3 male, average age 9.7 years) and site B (N = 5, 1 male, average age 11.6 years). For each participant, regardless of site, we acquire structural T1 weighted MRI (T1w MRI), resting state functional MRI (fMRI), diffusion tensor MRI (DTI), and magnetic resonance spectroscopy (MRS). At site A, all images were acquired on a 3T Philips Achieva scanner (Amsterdam, the Netherlands), and at site B, all images were acquired on a 3T Siemens Trio TIM scanner (Erlangen, Germany).
The T1w MRI data were acquired at 1.0mm isotropic resolution with an echo time (TE) and repetition time (TR) of TE/TR = 2.9ms/6310ms at site A and 3.5ms/2500ms at site B. The fMRI data were acquired at resting state with TE/TR = 30ms/2000ms at 2.25x2.25x2.4mm (sagittal, coronal, axial) resolution at site A and TE/TR = 30ms/2000ms at 2.51mm isotropic resolution at site B. Between 100 and 240 volumes were acquired for each participant. Two participants, one at each site, did not complete this acquisition. The DTI data were acquired with multishell acquisitions at b = 1000 s/mm2 (32 directions at site A with TE/TR = 70ms/4600ms at 1.9x1.9x2.0mm resolution and 30 directions at site B with TE/TR = 97.4ms/3800ms at 2.0mm isotropic resolution) and b = 2000 s/mm2 (32 directions at site A with TE/TR = 70ms/4600ms at 1.9x1.9x2.0mm resolution and 30 directions at site B with TE/TR = 97.4ms/3800ms at 2.0mm isotropic resolution) phase-encoded in the anterior-to-posterior direction with associated b = 0 s/mm2 non-diffusion weighted volumes. Additional b = 0 s/mm2 non-diffusion weighted volumes phase encoded in the opposite direction were acquired as well at each site for susceptibility-induced distortion correction [9], [10]. One participant at site A did not complete this acquisition. The MRS data were acquired with a 2-dimensional point resolved spectroscopy (PRESS) sequence in an axial slice at the level of the basal ganglia. Images at site A were acquired with TE = 144ms at 9.58x9.58x15mm resolution. Images at site B were acquired with TE = 135ms at 12.5x12.5x10mm resolution.
2.2. Multimodal image processing
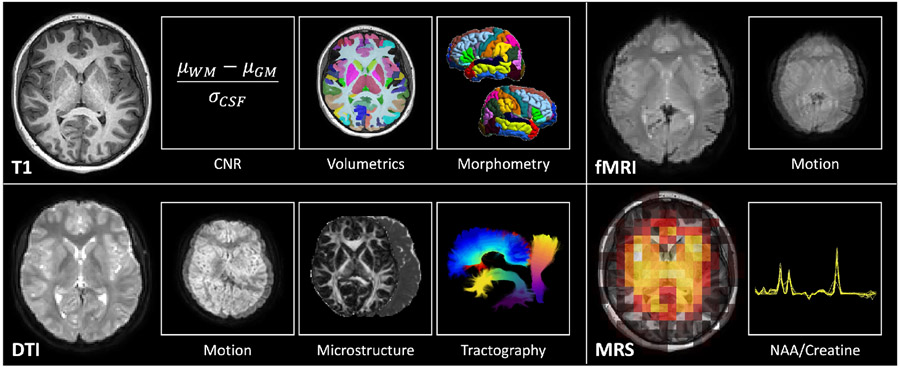
For each T1w MRI, we compute gray matter (GM) volumes in 34 frontal regions, 14 parietal regions, 22 temporal regions, 16 occipital regions, and 19 deep regions with the spatially localized atlas network tiles (SLANT) deep learning framework [11]. During this process, SLANT also performs a whole brain, white matter (WM), and ventricle segmentation. The volumes of each GM region are subsequently normalized to the intracranial volume. Next, we compute the local gyrification index (LGI) in the same frontal, parietal, temporal, and occipital cortical regions [12]-[15]. For each resting state fMRI, we compute the median voxel-wise intervolume displacement with statistical parametric mapping (SPM) [16]. For each DTI, we preprocess the images with the PreQual pipeline [10], extract the b = 1000 s/mm2 volumes, and fit a tensor model to compute scalar diffusion microstructural fractional anisotropy (FA) and mean diffusivity (MD) maps [17]. Next, we extract the b = 2000 s/mm2 volumes and perform probabilistic tractography [18], generating a whole brain tractogram, which is then subsequently parcellated into 32 white matter bundles [19], and compute the average FA and MD along each of the bundles [20], [21]. For each MRS, we fit the spectra in each voxel with LCModel [22], computing the ratios of the N-acetylaspartate (NAA) peaks to the creatine (Cr) peaks. Last, we compute the median voxel-wise NAA/Cr ratio in the middle ninth of the slice. These operations are summarized in Figure 1.
Figure 1.
Multimodal neuroimaging of pediatric T1D at each site. We acquire T1w MRI, DTI, resting state fMRI, and MRS. For T1w MRI, we characterize image CNR, gray matter volumetrics, and cortical morphometry. For DTI, we characterize intervolume motion and diffusion microstructure along white matter bundles identified with tractography. For fMRI, we characterize intervolume motion. For MRS, we characterize the NAA/Cr ratio in an axial slice through the basal ganglia.
2.3. T1 weighted MRI
To characterize T1w MRI quality, we evaluate the contrast to noise ratio (CNR) of each image. With the SLANT segmentations of GM, WM, and the ventricles, we compute the average signal in GM, μGM, and WM, μWM, as well as the standard deviation of the cerebrospinal fluid (CSF) signal, σCSF. CNR is then computed as:
| (1) |
We compare the distributions of CNR in T1w MRI between participants at both sites with the Wilcoxon rank-sum test at 0.05 significance.
Next, we compare the normalized GM volumes quantified with SLANT between the two sites for the regions in each cortical lobe and the deep GM regions. To do so, we model the GM volumes for each set of regions with a linear mixed effects model at 0.05 significance, considering site and region as fixed effects and participants as a random effect. We plot the distribution of regional median GM volumes across participants for each site and report the p-value for the site effect with Bonferroni correction for five comparisons (four lobes and deep GM). Additionally, we compute the coefficient of variation (CoV) for each region across participants at each site to better quantify and compare within-site variability. We compare the distribution of regional CoV for each site and evaluate for statistical differences with the Wilcoxon rank-sum test at 0.05 significance with Bonferroni correction for the same five comparisons.
Last, we compare the regional cortical LGIs in the four lobes in the same manner. We model the LGI for each set of regions with a linear mixed effects model in the same configuration at 0.05 significance, plot the distribution of regional median LGI, and report the p-value for the site effect with Bonferroni correction for four comparisons, one for each lobe. We repeat the CoV analysis between sites, comparing the distribution of regional CoV at each site with the Wilcoxon rank-sum test at 0.05 significance with Bonferroni correction for the same four comparisons.
2.4. Resting state functional MRI
To characterize resting state fMRI quality, we evaluate the intervolume motion. We average the median voxel-wise intervolume displacement computed with SPM across volumes for each participant. We then compare the distribution of average intervolume displacement between sites. We evaluate for statistical differences with the Wilcoxon rank-sum test at 0.05 significance.
2.5. Diffusion tensor MRI
To characterize DTI quality, we first evaluate intervolume motion. Similarly to fMRI, we average across volumes the average voxel-wise intervolume displacement computed with the eddy preprocessing tool in the PreQual preprocessing pipeline [10], [23]. We then compare the distribution of average intervolume displacement between sites with the Wilcoxon rank-sum test at 0.05 significance.
Second, we evaluate diffusion microstructure measured along the 32 white matter bundles virtually dissected with tractography. Similar to the GM volumes and LGI in T1w MRI, we model the average FA and MD along each of the bundles with a linear mixed effects model at 0.05 significance, considering site and bundle as fixed effects and participants as a random effect, and plot the distribution of bundle-wise median FA and MD across participants for each site and report the p-value for the site effect. We perform a similar CoV analysis to that described for GM volume and LGI. We compute the CoV for each bundle across participants and compare the distributions of bundle-wise CoV for each site with the Wilcoxon rank-sum test at 0.05 significance. Last, we utilize the ComBat tool to harmonize the bundle-wise microstructural scalars between sites, and repeat the bundle-wise FA, MD, and CoV analyses described for the harmonized data [24].
2.6. Magnetic resonance spectroscopy
To characterize MRS, we evaluate the median NAA/Cr ratio in the middle ninth of an axial slice through the basal ganglia for each participant. We compare the distributions at each site with a Wilcoxon rank-sum test at 0.05 significance. We perform harmonization between sites to account for TE differences with a phantom study at site A. We scan a General Electric Braino phantom (Boston, USA) with the site A protocol at a TE of both 144ms (matching site A) and 135ms (matching site B). We subsequently quantify the median scale factor between the NAA/Cr ratios in the 135ms scan and the 144ms scan in the middle of the phantom. We multiply this scale factor to the NAA/Cr ratios measured at site A to harmonize the TE to that at site B. We compare the harmonized distributions between sites in the same manner.
2.7. Sample size calculations
To investigate the feasibility of a large-scale multimodal, multisite neuroimaging study of T1D, we investigate the sample sizes needed for each modality to identify a 5% difference between a group of participants with T1D and a group of controls. We model the imaging measures as continuous response variables from independent control and experimental participants with T1D at a ratio of 1 control per case with 0.9 power and 0.05 significance. For each modality, we assume the responses are normally distributed and that the true difference between the control and experimental means is 5% of the mean values with a standard deviation computed by multiplying the mean values by the associated CoV. We compute mean values and CoV for this analysis across all participants from this pilot study.
Specifically, for T1w MRI, for each region we compute the average GM volume and LGI and the corresponding CoV across our entire pilot study population. We do the same for bundle-wise FA and MD in DTI after harmonization and for median NAA/Cr in the middle ninth of the slice for MRS after harmonization. Subsequently, for T1w MRI and DTI, we compute the sample size required for each region or bundle to identify a 2% through 10% difference between groups. We obtain a distribution of sample sizes across regions for each percent difference. We do the same for MRS, but compute only one sample size estimate per percent difference as there are no regions involved. Last, we plot these estimates and visually estimate the sample size needed to resolve, on average, a 5% difference between groups across all modalities.
3. RESULTS
3.1. T1 weighted MRI
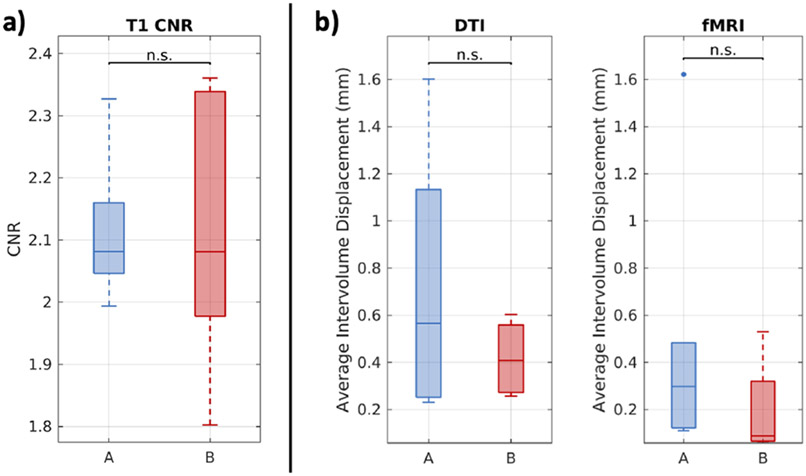
The distributions of CNR at both sites are reported in Figure 2a. We find that the CNR between sites exhibits no statistically significant difference. We find that participants at both sites exhibit a difference in signal between GM and WM that is approximately double the standard deviation of CSF signal in the ventricles. We find the spread of CNR at site B is larger, but we do not find the absolute CNR values of 1.8 to 2.4 to be unacceptable for further analysis.
Figure 2.
Multimodal image contrast and motion. (a) T1w MRI from the two sites exhibit comparable CNR. Statistical significance was evaluated with the Wilcoxon rank-sum test (p < 0.05). (b) With scans acquired around 2mm resolution, on average, participants experience subvoxel intervolume movement in both DTI and resting state fMRI at both sites. Statistical significance was evaluated with the Wilcoxon rank-sum test (p < 0.05).
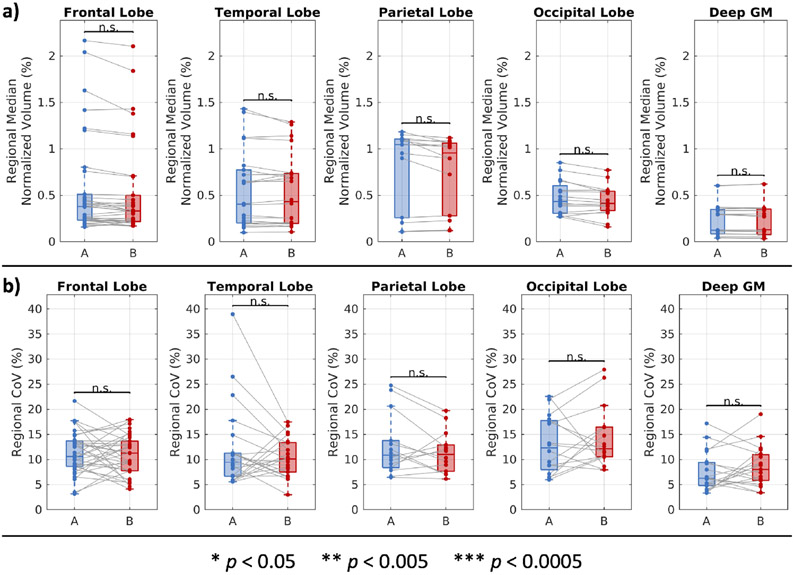
The distributions of regional normalized GM volume and associated CoVs are reported in Figure 3. We find that the volumes exhibit no statistically significant difference between sites across regions in the four cortical lobes and deep GM. We find the same for the CoV of those regions and that, across all four lobes and deep GM, the median CoV at each site range from 5 to 15%.
Figure 3.
Regional normalized GM volumes at two sites. (a) On a regional basis, GM volumes are comparable between sites. Statistical significance was evaluated with a linear mixed effects model with site and region as fixed effects and participants as a random effect. The p-values for the site effects are reported with Bonferroni correction for 5 comparisons. (b) On a regional basis, the CoV of measurements in the same areas are comparable between sites. Statistical significance was evaluated with the Wilcoxon rank-sum test with Bonferroni correction for 5 comparisons.
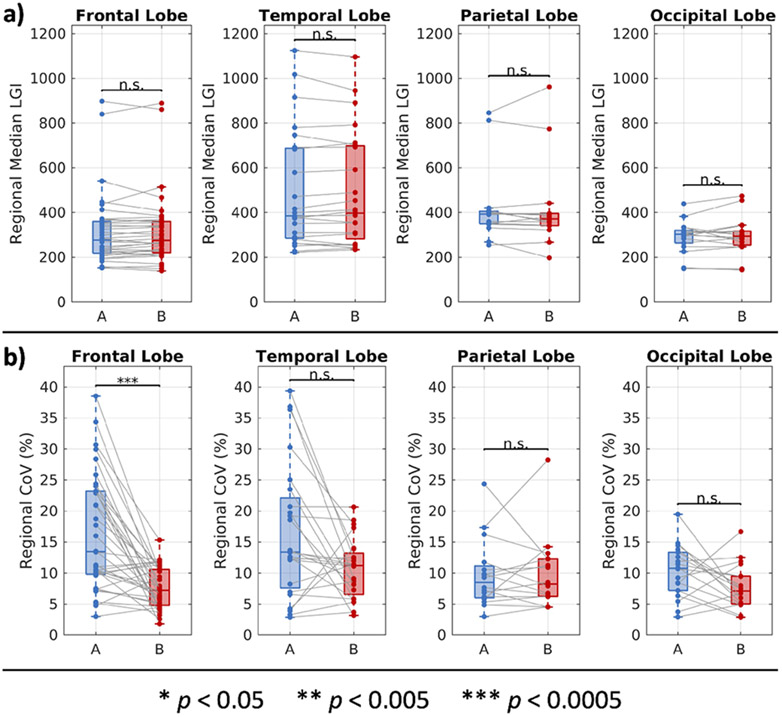
The distributions of regional LGI and associated CoVs are reported in Figure 4. We find that the LGI estimates exhibit no statistically significant differences between sites across the four lobes. We find the same for CoV in the temporal parietal and occipital lobes. For CoV in the frontal lobe, we find that, despite the statistically significant differences, the results still place an estimate of the central CoV between 5 and 15%, in line with the results from the other lobes.
Figure 4.
Regional measurements of local gyrification at two sites. (a) On a regional basis, cortical gyrification is comparable between sites. Statistical significance was evaluated with a linear mixed effects model with site and region as fixed effects and participants as a random effect. The p-values for the site effects are reported with Bonferroni correction for four comparisons. (b) On a regional basis, the CoV of measurements in the same regions are comparable between sites. The statistically significant differences in frontal lobe regional CoV still primarily fall within the same 5-15% CoV as the other lobes. Statistical significance was evaluated with the Wilcoxon rank-sum test with Bonferroni correction for four comparisons.
3.2. Resting state functional MRI
We find that the average fMRI intervolume movement trends downward between site A and site B, but that the maximum average intervolume displacement experienced by participants at both sites was on the subvoxel level for acquisitions acquired around 2mm resolution (Figure 2b). This indicates to us that movement in pediatric T1D participants during resting state fMRI in this pilot study was low.
3.3. Diffusion tensor MRI
As with fMRI, we find that the average DTI intervolume movement trends downward between site A and site B, but that the maximum average intervolume displacement is on the subvoxel level (Figure 2b). This indicates to us that the pediatric T1D participants in this pilot study were able to undergo multishell DTI with low amounts of motion.
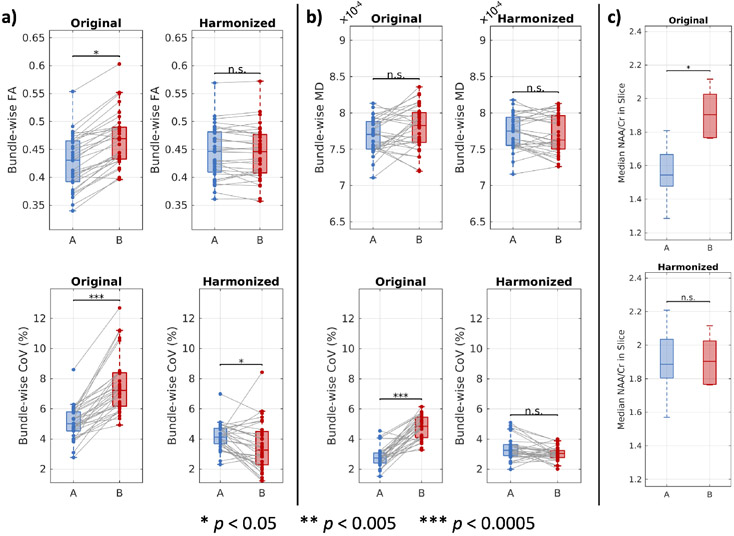
The distributions of bundle-wise FA and MD and the associated CoVs are reported in Figure 5. We find that, without harmonization, bundle-wise FA exhibited statistically significant differences between sites that was eliminated with harmonization (Figure 5a). On the other hand, we find that bundle-wise MD did not exhibit statistically significant differences between sites both without and with harmonization (Figure 5b). We also find that harmonization reduces differences in CoV between sites and that, overall, the CoV estimates fall between 2% and 5% (Figure 5a and 5b).
Figure 5.
Bundle-wise measurements of diffusion microstructure and NAA to creatine ratios in a slice through the basal ganglia at two sites before and after harmonization. (a) On a bundle basis, average FA and the associated CoV are not comparable between sites without harmonization. After harmonization, the FA values are comparable, and the differences in CoV between sites are reduced and small in absolute value. (b) On a bundle basis without harmonization, average MD is comparable between sites, but the associated CoV is not. After harmonization, the differences in CoV between sites are reduced and small in absolute value. (a and b) Harmonization was performed with the ComBat method. Statistical significance for the microstructural scalars was evaluated with a linear mixed effects model with site and bundle as fixed effects and participants as a random effect. The p-values for the site effects are reported. Statistical significance for the CoV estimates was evaluated with the Wilcoxon rank-sum test. (c) NAA/Cr ratios are not comparable between the sites without harmonization. Harmonization was performed with a phantom study at site A to correct for TE differences. Statistical significance was determined with the Wilcoxon rank-sum test (p < 0.05).
3.4. Magnetic resonance spectroscopy
The distributions of median NAA/Cr in the middle ninth of an axial slice through the basal ganglia among participants at each site are reported in Figure 5c. We find that before harmonization the ratios exhibit statistically significant differences between sites and that those differences are corrected after harmonization leveraging a phantom study at site A.
3.5. Sample size calculations
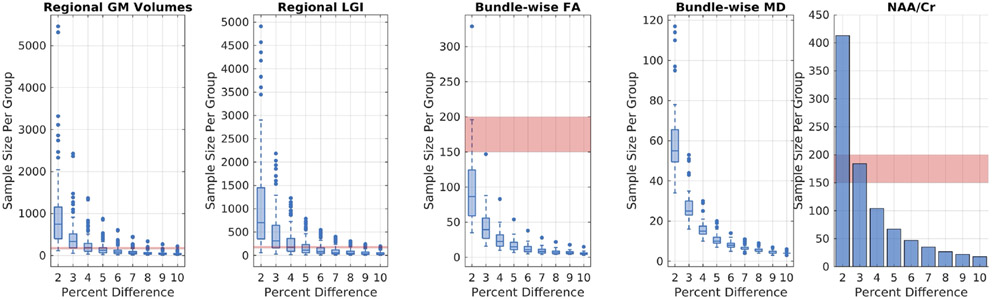
The results of the multimodal sample size computations are reported in Figure 6. We find that a sample of approximately 150-200 participants with T1D and the same number of healthy controls is needed to resolve a 5% difference between groups across all the modalities investigated presently at 0.9 power and 0.05 significance.
Figure 6.
Power analysis of multimodal, multisite imaging of pediatric T1D. At 0.9 power and 0.05 significance, a sample size of ~150-200 (red highlight) per group is necessary to detect a 5% difference, on average, in the pediatric T1D multimodal measures investigated presently.
4. DISCUSSION AND CONCLUSIONS
We present a pilot study for multisite, multimodal neuroimaging feasibility in pediatric T1D to better elucidate its complex, multifactorial neuroanatomical effects. We find the T1-based measures to be comparable between sites without harmonization and find the DTI and MRS-based measures to be comparable after harmonization. We find that acquiring both DTI and fMRI is feasible with low motion artifact. We find 5-15% coefficient of variation within sites for almost all measures, suggesting a sample size of approximately 150-200 participants with T1D and the same number of controls are needed on average to detect a 5% difference across these modalities at 0.9 power. As such, we conclude that multimodal neuroimaging of pediatric T1D is feasible at multiple sites with low motion artifact, taking into account harmonization of DTI and MRS.
Data strongly implicate diabetic ketoacidosis (DKA) and hyperglycemia in causing cognitive declines in children and adults with T1D that may worsen over time [25], [26]. Declines in cognitive function may lead to suboptimal T1D management, resulting in a vicious cycle that increases risk for poor glycemic control, life-threatening complications, and further cognitive declines [27]. The mechanisms underlying neural injury are unclear, and a longitudinal cohort study of children with T1D is necessary to characterize associations between T1D-related factors and neurocognitive outcomes. Multimodal neuroimaging with consideration of site and scanner effects in a large cohort will identify macrostructural, microstructural, inflammatory, and functional neuroimaging correlates of impaired cognition and will assist in identifying potentially modifiable mechanisms that may ameliorate cognitive declines in children with T1D.
The primary limitation of this pilot study is the small sample size. Due to this, the comparisons of distributions across participants between sites without further stratification are likely underpowered. This includes the CNR, motion, and MRS analyses. Additionally, since the regions within one participant’s brain are highly correlated, regional effects had to be considered when comparing distributions of values across regions. With a small sample size, this can impact the ability of the linear mixed effects model to resolve the site effect. However, it is reassuring in both cases that there are no immediate trends suggesting large differences between sites.
On a related note, because the regions are highly correlated, they are likely unable to be considered fully independent samples. This indicates the independence assumption in the Wilcoxon rank-sum tests used to evaluate differences in CoV between sites may not hold. For this pilot study, we selected this approach to facilitate a more interpretable metric, like CoV, in order to perform subsequent sample size analyses. In the future, regional effects ought to be taken into consideration when analyzing within site variability.
We note that interscanner harmonization is an open problem in neuroimaging. For this pilot study, we selected a phantom-based correction for TE effects in MRS and the commonly used ComBat method for DTI for their interpretability and ease of implementation. However, future directions include leveraging other techniques for improved harmonization. For instance, the selected TE ought to be standardized from site to site for MRS. Additionally, for DTI, more advanced techniques, like harmonization of spherical harmonics-based features or deep learning-based methods, ought to be considered [28], [29].
Compared to the other imaging modalities, MRS suffers from low spatial resolution. For this pilot study, we approximately localized deep GM structures by utilizing the voxels in the middle ninth of the slice through the basal ganglia. However, this necessarily will also encompass signal from the ventricles and WM. Future directions include leveraging the SLANT segmentations to better separate specific regions of the brain for metabolite analysis.
Last, there are other processing techniques beyond those used here that can be investigated in context of multimodal imaging in T1D. These include applying distortion correction to fMRI prior to analysis and functional connectomics; other methods of segmenting GM from T1w MRI including multi-atlas approaches; investigation of other cortical morphometry measures like curvature, thickness, and shape; the investigation of multishell DTI micro structural scalars, like the neurite orientation dispersion and density imaging measures; and structural connectomics.
ACKNOWLEDGEMENTS
The authors thank Nowrin U. Chowdhury for a thorough read-through of this manuscript. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work was supported by the National Institutes of Health (NIH) under award numbers 1U34DK123895-01, U34DK123894-01, 1R01MH121620-01, and T32GM007347 and by the National Science Foundation under award number 2040462. This research was conducted with the support from the Intramural Research Program of the National Institute on Aging of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- [1].Brands AMA, Biessels GJ, De Haan EHF, Kappelle LJ, and Kessels RPC, “The effects of type 1 diabetes on cognitive performance: A meta-analysis,” Diabetes Care, vol. 28, no. 3, pp. 726–735, 2005, doi: 10.2337/diacare.28.3.726. [DOI] [PubMed] [Google Scholar]
- [2].Centers for Disease Control and Prevention, “National Diabetes Statistics Report: Estimates of Diabetes and its Burden in the United States,” 2014.
- [3].Mazaika PK et al. , “Variations in brain volume and growth in young children with type 1 diabetes,” Diabetes. vol. 65, no. 2, pp. 476–485, Feb. 2016, doi: 10.2337/db15-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wessels AM et al. , “Cognitive performance in type 1 diabetes patients is associated with cerebral white matter volume,” Diabetologia, vol. 50, no. 8, pp. 1763–1769, 2007, doi: 10.1007/s00125-007-0714-0. [DOI] [PubMed] [Google Scholar]
- [5].Antenor-Dorsey JAV, Shimony JS, and Hershey T, “Diffusion Tensor Imaging of the Brain in Type 1 Diabetes,” EMJ Diabetes, vol. 2, no. October, pp. 42–47, 2014. [Google Scholar]
- [6].Shukla V, Shakya AK, Perez-Pinzon MA, and Dave KR, “Cerebral ischemic damage in diabetes: An inflammatory perspective,” J. Neuroinflammation, vol. 14, no. 1, pp. 1–22, 2017, doi: 10.1186/s12974-016-0774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Muriach M, Flores-Bellver M, Romero FJ, and Barcia JM, “Diabetes and the brain: Oxidative stress, inflammation, and autophagy,” Oxid. Med. Cell. Longev, vol. 2014, 2014, doi: 10.1155/2014/102158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cai LY et al. , “MASiVar: Multisite, multiscanner, and multisubject acquisitions for studying variability in diffusion weighted MRI.” Magn. Reson. Med, no. March, pp. 1–17, 2021, doi: 10.1002/mrm.28926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Andersson JLR. Skare S, and Ashburner J, “How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging,” Neuroimage, vol. 20, no. 2, pp. 870–888, Oct. 2003, doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- [10].Cai LY et al. , “PreQual: An automated pipeline for integrated preprocessing and quality assurance of diffusion weighted MRI images,” Magn. Reson. Med, no. December 2020, pp. 1–15, 2021, doi: 10.1002/mrm.28678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huo Y et al. “3D whole brain segmentation using spatially localized atlas network tiles,” Neuroimage, vol. 194, no. November 2018, pp. 105–119, 2019, doi: 10.1016/j.neuroimage.2019.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lyu I, Kim SH, Girault JB, Gilmore JH, and Styner MA, “A cortical shape-adaptive approach to local gyrification index,” Med. Image Anal, vol. 48, pp. 244–258, 2018, doi: 10.1016/j.media.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lyu I, Kim SH, Woodward ND, Styner MA, and Landman BA, “TRACE: A Topological Graph Representation for Automatic Sulcal Curve Extraction,” IEEE Trans. Med. Imaging, vol. 37, no. 7, pp. 1653–1663, 2018, doi: 10.1109/TMI.2017.2787589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lyu I, Kang H, Woodward ND, Styner MA, and Landman BA, “Hierarchical spherical deformation for cortical surface registration,” Med. Image Anal, vol. 57, pp. 72–88, 2019, doi: 10.1016/j.media.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Parvathaneni P et al. “Cortical Surface Parcellation Using Spherical Convolutional Neural Networks,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2019, vol. 11766 LNCS, pp. 501–509, doi: 10.1007/978-3-030-32248-9_56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, and Nichols TE, Statistical parametric mapping: the analysis of functional brain images. Elsevier, 2011. [Google Scholar]
- [17].Westin CF, Peled S, Gudbjartsson H, Kikinis R, and Jolesz FA, “Geometrical Diffusion Measures for MRI from Tensor Basis Analysis,” Proc. 5th Annu. Meet. ISMRM. p. 1742, 1997. [Google Scholar]
- [18].Theaud G, Houde JC, Boré A, Rheault F, Morency F, and Descoteaux M, “TractoFlow: A robust, efficient and reproducible diffusion MRI pipeline leveraging Nextflow & Singularity,” Neuroimage, vol. 218, no. May, 2020, doi: 10.1016/j.neuroimage.2020.116889. [DOI] [PubMed] [Google Scholar]
- [19].Garyfallidis E et al. , “Recognition of white matter bundles using local and global streamline-based registration and clustering,” NeuroImage, vol. 170. Academic Press Inc., pp. 283–295, Apr. 15, 2018, doi: 10.1016/j.neuroimage.2017.07.015. [DOI] [PubMed] [Google Scholar]
- [20].Chandio BQ et al. “Bundle analytics, a computational framework for investigating the shapes and profiles of brain pathways across populations,” Sci. Rep, vol. 10, no. 1, pp. 1–18, 2020, doi: 10.1038/s41598-020-74054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yeatman JD, Dougherty RF, Myall NJ, Wandell BA, and Feldman HM. “Tract Profiles of White Matter Properties: Automating Fiber-Tract Quantification,” PLoS One, vol. 7, no. 11, p. 49790, 2012, doi: 10.1371/journal.pone.0049790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Provencher SW, “Estimation of metabolite concentrations from localized in vivo proton NMR spectra,” Magn. Reson. Med, vol. 30, no. 6, pp. 672–679, 1993, doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- [23].Andersson JLR and Sotiropoulos SN, “An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging,” Neuroimage, vol. 125, pp. 1063–1078, 2016, doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fortin JP et al. , “Harmonization of multi-site diffusion tensor imaging data,” Neuroimage, vol. 161, no. July, pp. 149–170, 2017, doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cameron FJ et al. , “Neurological consequences of diabetic ketoacidosis at initial presentation of type 1 diabetes in a prospective cohort study of children,” Diabetes Care, vol. 37, no. 6, pp. 1554–1562, 2014, doi: 10.2337/dc13-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ghetti S et al. , “Cognitive function following diabetic ketoacidosis in children with new-onset or previously diagnosed type 1 diabetes,” Diabetes Care, vol. 43, no. 11, pp. 2768–2775, 2020, doi: 10.2337/dc20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ryan CM, “Why is cognitive dysfunction associated with the development of diabetes early in life? The diathesis hypothesis,” Pediatr. Diabetes, vol. 7, no. 5, pp. 289–297, 2006, doi: 10.1111/j.1399-5448.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- [28].Nath V et al. , “Harmonizing 1.5T/3T diffusion weighted MRI through development of deep learning stabilized microarchitecture estimators,” no. March 2019, p. 23, 2019, doi: 10.1117/12.2512902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mirzaalian H et al. , “Inter-site and inter-scanner diffusion MRI data harmonization,” Neuroimage, vol. 135, pp. 311–323, 2016, doi: 10.1016/j.neuroimage.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]