Abstract
In lung cancer screening, estimation of future lung cancer risk is usually guided by demographics and smoking status. The role of constitutional profiles of human body, a.k.a. body habitus, is increasingly understood to be important, but has not been integrated into risk models. Chest low dose computed tomography (LDCT) is the standard imaging study in lung cancer screening, with the capability to discriminate differences in body composition and organ arrangement in the thorax. We hypothesize that the primary phenotypes identified using lung screening chest LDCT can form a representation of body habitus and add predictive power for lung cancer risk stratification. In this pilot study, we evaluated the feasibility of body habitus image-based phenotyping on a large lung screening LDCT dataset. A thoracic imaging manifold was estimated based on an intensity-based pairwise (dis)similarity metric for pairs of spatial normalized chest LDCT images. We applied the hierarchical clustering method on this manifold to identify the primary phenotypes. Body habitus features of each identified phenotype were evaluated and associated with future lung cancer risk using time-to-event analysis. We evaluated the method on the baseline LDCT scans of 1,200 male subjects sampled from National Lung Screening Trial. Five primary phenotypes were identified, which were associated with highly distinguishable clinical and body habitus features. Time-to-event analysis against future lung cancer incidences showed two of the five identified phenotypes were associated with elevated future lung cancer risks (HR=1.61, 95% CI = [1.08, 2.38], p=0.019; HR=1.67, 95% CI = [0.98, 2.86], p=0.057). These results indicated that it is feasible to capture the body habitus by image-base phenotyping using lung screening LDCT and the learned body habitus representation can potentially add value for future lung cancer risk stratification.
Keywords: Body Habitus, Image-based Phenotyping, Lung Cancer Screening
1. INTRODUCTION
Body habitus refers to the physical and constitutional characteristics of human body, which includes the body composition profiles (e.g., muscle and adipose tissues) and organ arrangement [1]. In addition to a reflection of genetics, body habitus can depict the underlying macro conditions of the body, including nutrition level [2], metabolic status [3], and hormone condition [4], which influence the strength of immune system [5, 6]. Abnormal body habitus can serve as an indicator of progression of certain diseases. For instance, depending on evolvement of the disease, the ‘pink puffer’ and ‘blue bloater’ are two common phenotypes of chronic obstructive pulmonary disease (COPD) [7]. Under the context of lung cancer screening, body habitus is commonly assessed by body mass index (BMI), which is inversely associated with the risk of future lung cancer incidence and has been included as a predictor variable in existing risk estimation models, e.g., PLCOm2012 [8]. However, BMI has been long criticized for its lack of capability to differentiate muscle, fat, and skeletal mass, or depict the detailed organ arrangement, implying that individuals with similar BMI may have dramatically different body habitus [9–11]. All indicate the necessity to study the representation for body habitus and its predictive power in lung cancer risk stratification.
Chest low dose computed tomography (LDCT) is the standard practice in lung cancer screening [12]. In addition to the lung parenchyma, these thoracic computed tomography (CT) images can provide high resolution depictions of other anatomical structures in the thorax, including body compositions, bones, cardiovascular system, and overall thoracic cavity morphology. This makes chest LDCT an ideal modality to study the representation for body habitus in the context of lung cancer risk prediction. Figure 1 shows two examples with dramatically different body habitus, which is clearly depicted by lung screening LDCT. Current methods for body habitus evaluation using CT images are mainly based on quantitative measurement of certain organs. For example, the body composition profiles were depicted by the measured cross-sectional areas of muscle and adipose tissue on axial CT slices selected at certain landmarks [13]. The thoracic cavity morphology was described by the dimensions of lung regions on given anatomical directions [14]. However, these quantitative measures can only capture the anatomical information from certain aspects and lack the capability to utilize the entirety of the rich anatomical information encoded in the imaging data.
Figure 1.
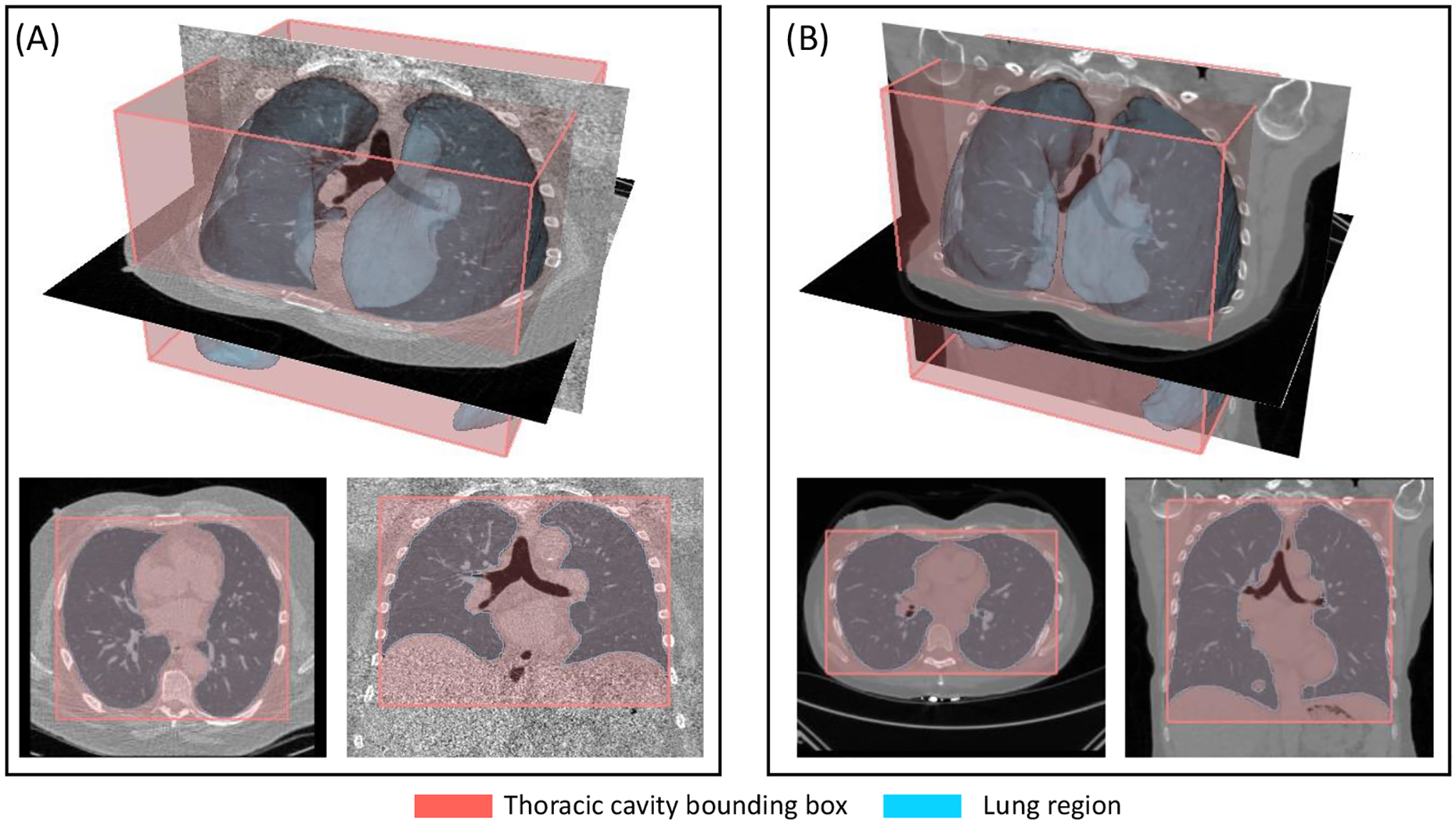
Lung screening LDCT is capable of distinguishing subjects with different thoracic body habitus. For the two examples, the lung masks and thoracic cavity bounding boxes are overlayed with axial slices at T8 vertebral level and coronal slices at the tracheal bifurcation. The overlap is displayed in both 3D (top row) and planar (bottom row) views. (A) shows a subject with barrel chest and elevated diaphragm position, indicated by the increased ratio in anterior-posterior and transverse dimensions, and decreased ratio between inferior-superior and transverse dimensions of thoracic cage. (B) shows a subject with flattened thoracic cage, with lowered diaphragm position.
In this study, we considered the unsupervised image-based phenotyping approach that captured the body habitus subpopulations using the chest LDCT images of large-scale lung screening cohort. We followed the method described in [15] to approximate the anatomical manifold by defining an intensity-based pairwise (dis)similarity measurement for spatially normalized chest LDCT images, which was followed by hierarchical agglomerative clustering analysis to identify similar groups. The differences in identified phenotypes were characterized using demographic information, thoracic cavity morphology, body composition, emphysema involvement, and lung cancer incidence rate. In addition, we studied the association of identified phenotypes with future lung cancer risk with time-to-event analysis. Some preliminary results of this work have been presented in prior conference proceedings [16, 17]. In this paper, we include more detailed description for the technique, and additional materials for the characterization of the identified phenotypes.
2. METHOD
In this section, we introduce the technical details of this study. This includes the preprocess of the lung screening LDCT scans, the build of imaging manifold, clustering analysis, and evaluate method to characterize the clustering results. The overall study design is given in Figure 2.
Figure 2.
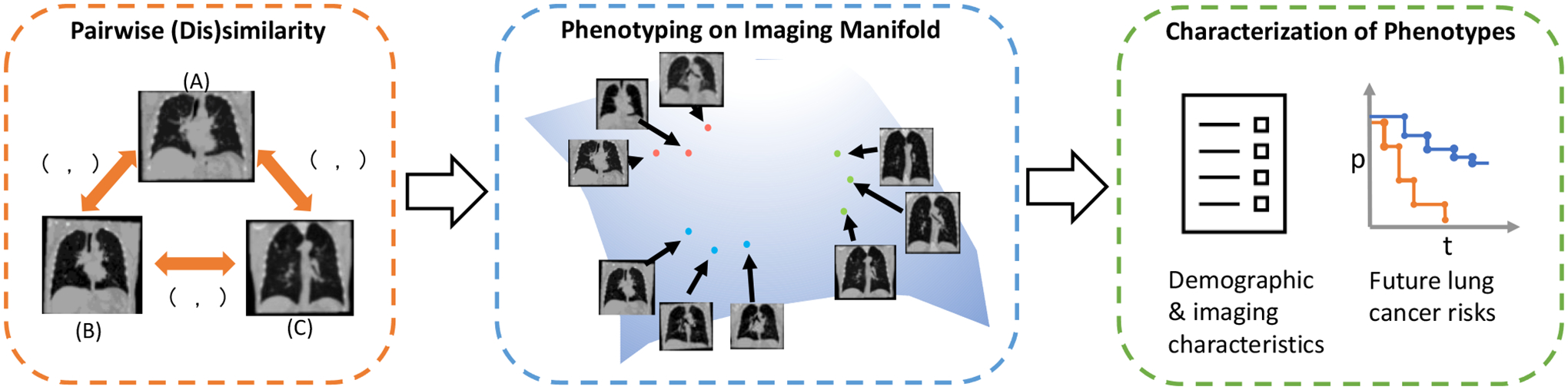
The overall design of this study. A pairwise (dis)similarity relationship is defined to approximate the thoracic imaging manifold. Phenotyping is conducted by applying hierarchical clustering algorithm on the approximated imaging manifold. We characterize the identified phenotypes for the demographic and imaging properties and the association with future lung cancer risk.
2.1. Image Pre-processing
The chest LDCT scans were converted into Hounsfield Units (HU). We first identified the specific regions in the image, which included the lung masks, using the published pretrained model given in [18], and the body masks, using a morphology-based method developed in [19]. The extraneous information like the scan tables and clothes were removed. To further normalize the images and reduce the variations introduced by different body positioning, the processed images were rigidly registered to a common reference space using the NiftyReg toolbox [20].
2.2. Imaging Manifold of Lung Screening LDCT
Imaging manifold provides an effective approach to reason and describe the anatomical variations in medical images [21–23]. The images can be regarded as points in the abstract manifold, with similar cases grouped closely and dissimilar cases arranged more far apart. With the high complexities and variations of anatomical structures in the thoracic space, explicit modeling of the anatomical manifold is almost infeasible. However, the structure of the anatomical manifold can be depicted empirically given the pairwise (dis)similarity between cases in the study cohort. Thus, the problem of defining the anatomical manifold can be simplified as formulating the (dis)similarity measurement between pairs of cases.
In this work, we followed the validated practice described in [15] and formulated the (dis)similarity measurement as intensity-based mean absolute difference (MAD) between two spatially normalized LDCT scans. The invalid regions and the regions outside the body were excluded from the MAD calculation, which mitigated the unrelated variations caused by FOV differences. The MAD (dis)similarity between two spatially normalized images A and B was given by
| (1) |
where R(·) refers to the set of pixels inside the body region of given image.
2.3. Cluster Analysis
Data-driven phenotype discovery is an effective technique to characterize the structure of a study cohort and expose unknown covariance in relation to certain disease of interest [24, 25]. Instead of using structured clinical features, we employed the anatomical manifold of lung screening LDCT to identify the homogeneous subgroups of the population. Like [15], we split the study cohort into phenotype groups using the intensity-based Hierarchical Agglomerative Clustering (HAC) method. Starting from single element clusters, the method iteratively merged closest cluster pairs until a single cluster status was reached. The pairwise similarity measurement was given by the intensity-based MAD. For the distance definition between clusters, we employed the complete-linkage strategy. This guaranteed the compactness of the resulting clusters as the intra-cluster pairs always have higher similarities than the inter-cluster pairs. The optimal number of clusters was identified by the elbow location of Residual Dissimilarity (RD) [15] with definition
| (2) |
where n is the number of clusters and represents the ith cluster. μ and σ refer to the mean and standard deviation of in-group pairwise (dis)similarity metric.
2.4. Result Characterization
To characterize the identified phenotypes, we retrieved multiple quantitative measurements from the lung screening LDCT image, including:
Thoracic cavity morphology. We measured the lung volume and the dimensions of the lung in transverse (T), anterior-posterior (AP), and inferior-superior (IS) directions in the spatially normalized space. These dimensions and the ratios between them can form an assessment for morphology of thoracic cavity.
Body composition. We applied the multi-level body composition assessment tool developed in [13]. The cross-sectional areas of subcutaneous adipose tissue (SAT) and skeletal muscle tissue (SMT) were measured on the axial slices at the fifth, eighth, and 10th vertebral locations. We also calculated the ratio between SAT and SMT.
Emphysema severity. The evolvement of emphysema was characterized by the percentage of lung with intensity less than −950 HU following the practice in [26].
In addition to these measurements, the identified phenotypes were characterized by demographics, smoking status, and lung cancer incidences. We assessed the significance across identified clusters using the Kruskall-Wallis test [27] for continuous variables and z-test for the proportional variables. We studied the time to lung cancer diagnosis for each cluster using the Kaplan-Meier (KM) estimation [28] and applied the log-rank test to assess for a difference in survival curves between the clusters. To study the effect of each cluster on future lung cancer risk, we used the Cox proportional hazards model [29] with the cluster with lowest index as reference. For all analyses, we specified an a priori significance threshold of 0.05 for each of our analyses. As this analysis was hypothesis generating, we did not adjust for multiple comparisons.
3. EXPERIMENTS AND RESULTS
3.1. Lung screening LDCT dataset
The study was conducted on a cohort selected from the CT arm of the National Lung Screening Trial (NLST) [30]. All image files and the patient specific information were obtained from the National Cancer Institute Cancer Data Access System (https://cdas.cancer.gov/nlst/). In this pilot study, we only considered male subjects in order to simplify the body habitus variation. Specifically, 1,200 male subjects were included, with 200 cases with lung cancer incidence during follow-up period. Both the follow-up time of non-cases and the time-to-diagnosis of lung cancer cases were available from the dataset repository. A characterization of this cohort is given in Table 1.
Table 1.
Characteristics of the NLST subset.
| Characteristic | Value |
|---|---|
| No. of subject | 1,200 |
| Age (y) | 62.5 ± 5.2 |
| BMI (kg/m2) | 27.9 ± 4.4 |
| Packyear | 62.1± 28.8 |
| Current smoker | 573 (47.8%) |
| Follow-up period (y) | 5.7 ± 2.0 |
| Lung cancer incidence | 200 (16.7%) |
In NLST, each subject had at most three longitudinal LDCT scans. Only the baseline scans were included in this study. The image preprocess and spatial normalization procedure described in Section 2.1 was applied on each scan. We reviewed each case and excluded those with non-standard body positioning, missing part of lung in field-of-view, or registration failure. This review process filtered out 14 problematic cases.
3.2. Image-based Phenotyping
We calculated the pairwise MAD (dis)similarity for the remaining 1,186 scans following Section 2.2, which was followed by the HAC analysis described in Section 2.3. Five primary clusters were identified following this procedure. To visualize the anatomical manifold, we employed the Uniform Manifold Approximation and Projection (UMAP) method [31]. UMAP seeks to learn the manifold structure of the data and find a low dimensional embedding, e.g., in 2-dimensional (2D) space, that preserves the essential topological structure of the manifold. UMAP can operate on a pre-defined distance matrix. For this study, we formulated this distance matrix using the pairwise MAD (dis)similarity. The resulting 2D visualization with clustering labels is shown in Figure 3 (a). We also obtained the average intensity maps for each identified cluster, which are demonstrated in Figure 3 (b). Systematic body habitus differences across clusters can be identified on these intensity maps, indicating the effectiveness of the image-base phenotyping approach.
Figure 3.
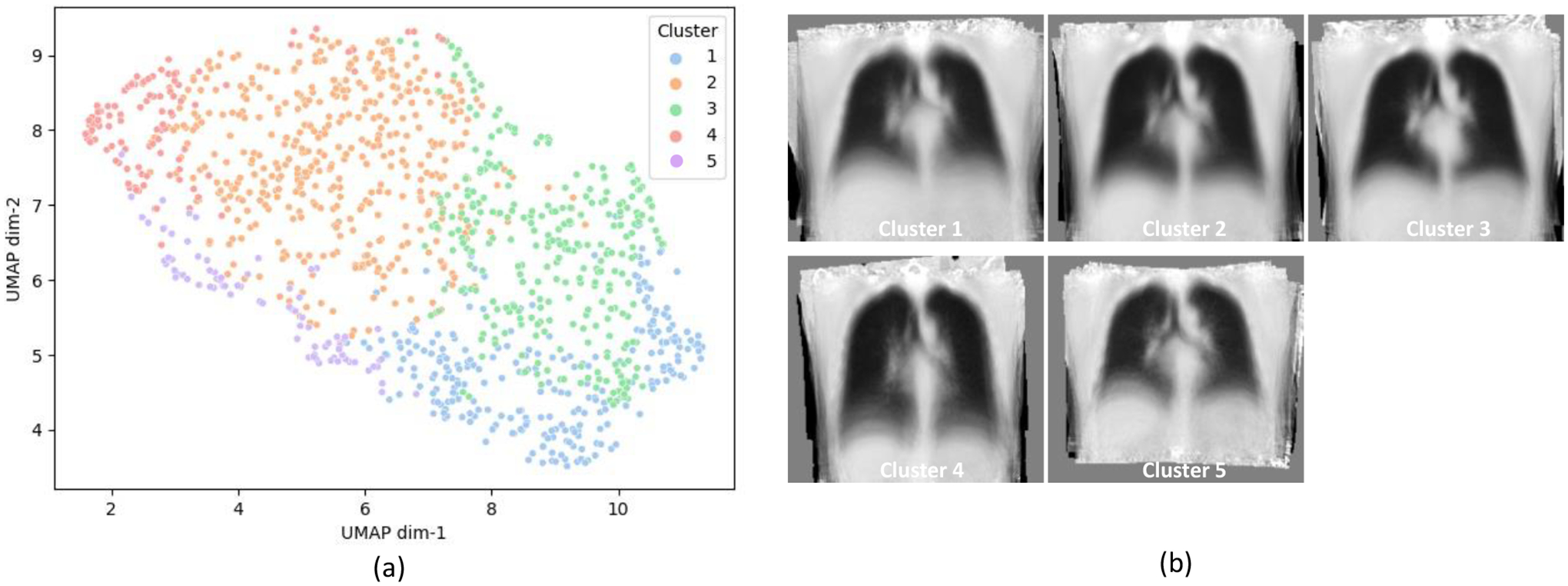
The algorithm described in Section 2 estimates five clusters. These clusters represent the primary phenotypes identified on the imaging manifold. (a) shows the UMAP display of the sample data distribution in imaging manifold with cluster labels. (b) shows the coronal views at tracheal bifurcation location of averaged intensity maps in the common space for each identified phenotype.
3.3. Characterization of Identified Phenotypes
We obtained the imaging measurements as described in Section 2.4, including descriptions for thoracic cavity morphology, body composition measurements, and per cent emphysema for characterization of emphysema evolvement. The statistics of these quantitative characteristics, together with demographics, smoking status, and lung cancer incidences across identified phenotypes, are given in Table 2. DTT, DTAP, and DTIS represent the lung dimensions measured in transverse, anterior-posterior, and inferior-superior directions, respectively. The KM estimation for lung cancer free rate and cluster-wise hazard ratios are given in Figure 4.
Table 2.
Characterization of identified phenotypes. All continuous properties are shown in mean values together with standard deviation (SD). The proportional properties are demonstrated in the number of incidences and percentage (%) in each phenotype group. The p-values are based on the equivalence test across phenotype groups, using Kruskall-Wallis test for continuous variables and z-test for the proportional variables.
| Characteristic | 1, N = 263 | 2, N = 439 | 3, N = 294 | 4, N = 109 | 5, N = 81 | p-value |
|---|---|---|---|---|---|---|
| Demographics & Smoking Status | ||||||
| Age in years (SD) | 62.23 (5.09) | 62.72 (5.32) | 62.21 (5.08) | 62.72 (5.24) | 62.53 (5.10) | p = 0.636 |
| BMI in kg/m2 (SD) | 32.15 (4.29) | 26.04 (2.84) | 28.91 (3.41) | 22.68 (2.08) | 27.98 (3.07) | p < 0.001 |
| Packyear (SD) | 64.21 (30.01) | 63.36 (31.01) | 62.43 (26.47) | 56.92 (23.14) | 54.02 (24.40) | p = 0.016 |
| No. current smoker (%) | 101 (38.4%) | 249 (56.7%) | 129 (43.9%) | 68 (62.4%) | 26 (32.1%) | p < 0.001 |
| Thoracic Cavity Morphology | ||||||
| Lung Volume in L (SD) | 6.13 (1.26) | 6.51 (1.10) | 6.87 (1.03) | 6.24 (1.17) | 4.74 (0.82) | p < 0.001 |
| DTT in mm (SD) | 278.16 (15.82) | 271.86 (15.03) | 277.75 (12.77) | 260.79 (15.66) | 263.38 (14.33) | p < 0.001 |
| DTAP in mm (SD) | 203.30 (12.01) | 183.47 (10.31) | 195.96 (10.49) | 167.84 (10.85) | 182.09 (11.37) | p < 0.001 |
| DTIS in mm (SD) | 215.15 (20.16) | 235.44 (17.98) | 231.95 (17.87) | 247.24 (19.98) | 205.63 (15.59) | p < 0.001 |
| DTT / DTAP (SD) | 1.37 (0.10) | 1.49 (0.11) | 1.42 (0.10) | 1.56 (0.12) | 1.45 (0.12) | p < 0.001 |
| DTIS / DTT (SD) | 0.77 (0.06) | 0.87 (0.06) | 0.84 (0.07) | 0.95 (0.08) | 0.78 (0.07) | p < 0.001 |
| Body Composition | ||||||
| SAT in cm2 (SD) | 529.90 (166.42) | 311.58 (119.68) | 410.32 (147.39) | 173.66 (96.03) | 387.22 (153.09) | p < 0.001 |
| SMT in cm2 (SD) | 460.73 (64.82) | 402.67 (55.70) | 443.21 (57.98) | 359.91 (52.51) | 419.18 (66.36) | p < 0.001 |
| SAT / SMT | 1.16 (0.38) | 0.79 (0.32) | 0.94 (0.35) | 0.50 (0.30) | 0.92 (0.33) | p < 0.001 |
| Disease | ||||||
| Percent Emphysema (SD) | 11.00 (8.76) | 13.26 (11.21) | 14.54 (11.18) | 15.82 (14.69) | 6.79 (7.43) | p < 0.001 |
| No. lung cancer incidence (%) | 34 (12.9%) | 88 (20.0%) | 41 (13.9%) | 22 (20.2%) | 10 (12.3%) | p = 0.041 |
Figure 4.
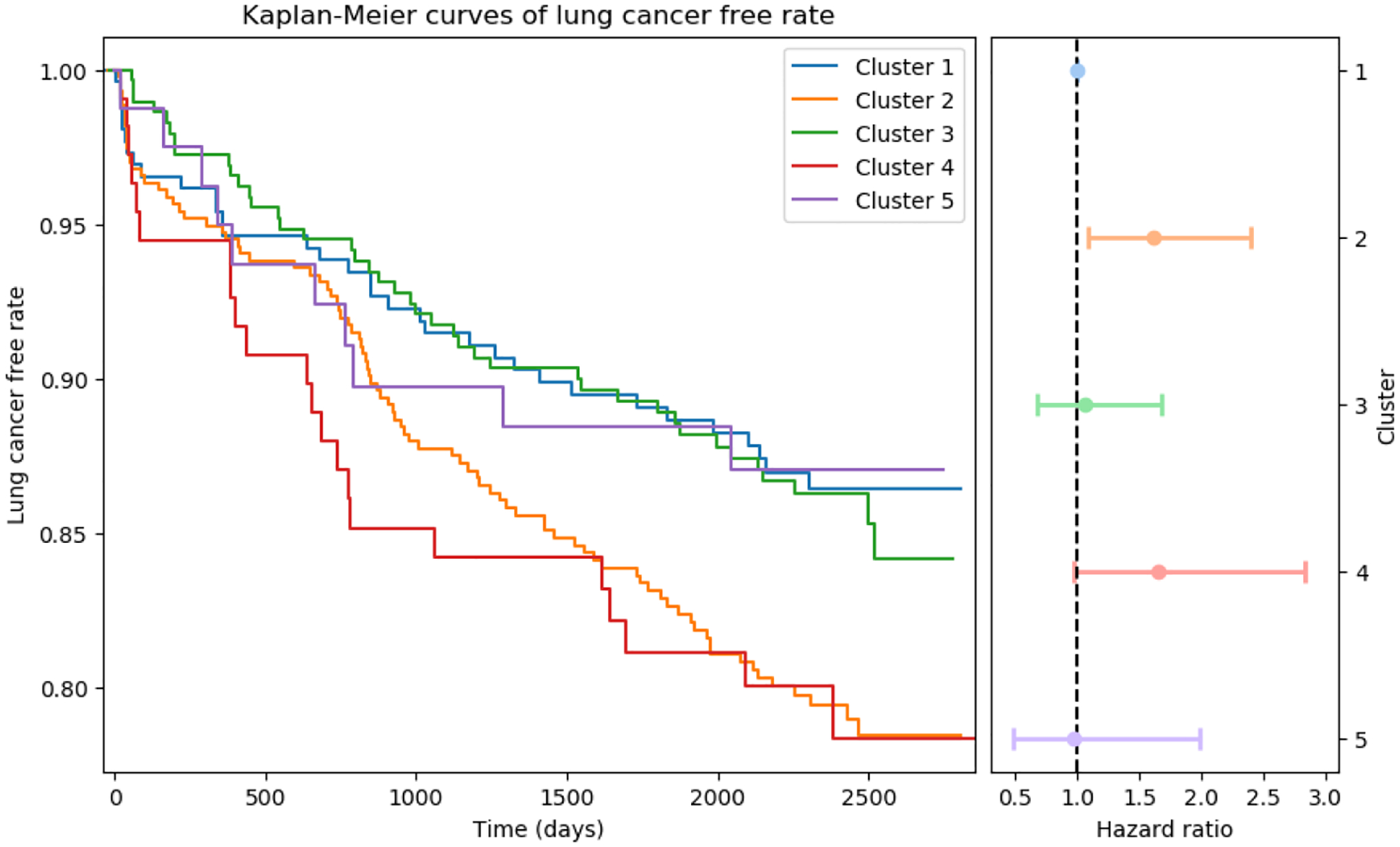
The left plot shows the Kaplan-Meier estimation of lung cancer free rate for each cluster, and the right plot shows the cluster-wise hazard ratio with 95% CI using cluster-1 as reference. Cluster-2 and cluster-4 are associated with elevated risk of lung cancer. These two phenotypes represent a cohort with increased percentage of current smoker, increased thoracic cavity dimension ratio in both anterior-posterior / transverse and inferior-superior / transverse directions, and decreased ratio between subcutaneous adipose tissue and skeletal muscle tissue.
The differences across the identified phenotypes were significance for all included variables except for age (Table 2). The characteristics of each phenotype were highly distinguishable and associated differently with future lung cancer risk. The detailed interpretations are given as follow:
Cluster-1. This cluster represented a phenotype with rounded (low DTT-DTAP ratio) and shallowed (low DTIS-DTT ratio) thoracic cavity. The estimated tissue mass for SAT and SMT were both higher than other phenotypes. The SAT was also account for a larger portion in body composition. The BMI for this group was the highest among the identified phenotypes. This phenotype was protective against future lung cancer risk (Table 2).
Cluster-2. The phenotype represented by cluster-2 was associated with more flattened thoracic cage shape (high DTT-DTAP ratio), with lowered diaphragm location (high DTIS-DTT ratio). The SAT accounted for a smaller portion of the body composition. BMI was also lower than averaged value (Table 1). The percentage of current smoker was significantly higher than the average level. This phenotype was associated with elevated risk of future lung cancer (HR = 1.61, 95% CI = [1.08, 2.38]; p = 0.019).
Cluster-3. This cluster represented a population with larger lung volume and slightly increased evolvement of emphysema. The other characteristics, including lung dimensions and dimension ratios, BMI, proportion of current smokers, and body composition profiles, all fell between the other two dominated clusters (cluster-1 and cluster-2), representing a ‘normal’ status. Even though emphysema was a known contributing factor for lung cancer risk, we did not observe increased future lung cancer incidence rate for this population.
Cluster-4. This cluster represented a smaller subpopulation with shared but strengthened characteristics of cluster-2, associated with extremely low BMI, higher rate of current smokers, more flattened thoracic cage, lower diaphragm position, decreased body composition mass, and decreased proportion of SAT in body composition. We observed an increased future lung cancer risk in this population (HR = 1.67, 95% CI = [0.98, 2.86]; p =0.057). Even though the p-value is above the priori significance threshold of 0.05, we believe this was caused by the limited size of the population considering the shared and strengthened characteristics against cluster-2.
Cluster-5. This cluster represented another smaller subpopulation but with unique characteristics against the other phenotypes. While the BMI, body composition profiles, and the horizontal ratio of thoracic cage were all fell in normal range, the phenotype was characterized by the decreased proportion of current smokers, decreased lung volume, elevated diaphragm position, and decreased evolvement of emphysema. We did not observe increase in lung cancer risk for this phenotype.
4. CONCLUSION
In this pilot study, we evaluated the feasibility for unsupervised image-based phenotyping using lung screening chest LDCT and its association with future lung cancer risk stratification. A pairwise intensity-based (dis)similarity metric was defined to approximate the anatomical manifold of chest LDCT. This was followed by hierarchical clustering analysis to identify the primary phenotypes. On an NLST subset, the algorithm identified five primary phenotypes. We characterized the identified phenotypes using demographics, smoking status, thoracic cavity morphology, body composition, emphysema evolvement, and lung cancer incidence rate. This characterization indicated that the approach could effectively capture clinically relevant phenotypes in the study population. With time-to-event analysis, we revealed that two of the identified phenotypes were associated with elevated risk of lung cancer. These results indicate that the unsupervised image-based phenotyping using lung screening LDCT is feasible and can potentially reveal novel imaging biomarkers with predictive power for future lung cancer risk stratification.
ACKNOWLEGEMENT
This research is supported or partly supported by the following awards: NSF CAREER 1452485; R01 EB017230; R01 CA253923; U01 CA196405 to Massion; Grant UL1 RR024975-01of the National Center for Research Resources and now Grant 2 UL1 TR000445-06 at the National Center for Advancing Translational Sciences; Martineau Innovation Fund Grant through the Vanderbilt-Ingram Cancer Center Thoracic Working Group; NCI Early Detection Research Network 2U01CA152662 to PPM; and IBM Fellowship.
REFERENCES
- [1].Zinn SL, [Body Size and Habitus] Butterworths, Boston, Chapter 137 (1990). [PubMed] [Google Scholar]
- [2].Thibault R, Genton L, and Pichard C, “Body composition: why, when and for who?,” Clin Nutr, 31(4), 435–47 (2012). [DOI] [PubMed] [Google Scholar]
- [3].Dulloo AG, Jacquet J, Solinas G et al. , “Body composition phenotypes in pathways to obesity and the metabolic syndrome,” Int J Obes (Lond), 34 Suppl 2, S4–17 (2010). [DOI] [PubMed] [Google Scholar]
- [4].Gates MA, Mekary RA, Chiu GR et al. , “Sex steroid hormone levels and body composition in men,” J Clin Endocrinol Metab, 98(6), 2442–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Odegaard JI, and Chawla A, “The immune system as a sensor of the metabolic state,” Immunity, 38(4), 644–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Taneja V, “Sex Hormones Determine Immune Response,” Front Immunol, 9, 1931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barnes PJ, Burney PG, Silverman EK et al. , “Chronic obstructive pulmonary disease,” Nat Rev Dis Primers, 1, 15076 (2015). [DOI] [PubMed] [Google Scholar]
- [8].Tammemagi MC, Katki HA, Hocking WG et al. , “Selection criteria for lung-cancer screening,” N Engl J Med, 368(8), 728–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cespedes Feliciano EM, Kroenke CH, and Caan BJ, “The Obesity Paradox in Cancer: How Important Is Muscle?,” Annu Rev Nutr, 38, 357–379 (2018). [DOI] [PubMed] [Google Scholar]
- [10].Bouchard C, “BMI, fat mass, abdominal adiposity and visceral fat: where is the ‘beef’?,” Int J Obes (Lond), 31(10), 1552–3 (2007). [DOI] [PubMed] [Google Scholar]
- [11].Gonzalez MC, Correia M, and Heymsfield SB, “A requiem for BMI in the clinical setting,” Curr Opin Clin Nutr Metab Care, 20(5), 314–321 (2017). [DOI] [PubMed] [Google Scholar]
- [12].Force USPST, Krist AH, Davidson KW et al. , “Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement,” JAMA, 325(10), 962–970 (2021). [DOI] [PubMed] [Google Scholar]
- [13].Xu K, Gao R, Tang Y et al. , [Extending the value of routine lung screening CT with quantitative body composition assessment] SPIE, MI: (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Montani D, Bertoletti L, Chaumais MC et al. , “Chronic thromboembolic pulmonary hypertension complicating long-term cyproterone acetate therapy,” Eur Respir Rev, 23(132), 260–3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jin Z, Udupa JK, and Torigian DA, “How many models/atlases are needed as priors for capturing anatomic population variations?,” Med Image Anal, 58, 101550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khan M, Xu K, Deppen S et al. , “Image-based Phenotyping for Risk Stratification in the National Lung Screening Trial.”
- [17].Khan M, Xu K, and Landman B, “Image-based Phenotyping for Improved Lung Cancer Risk Stratification.”
- [18].Hofmanninger J, Prayer F, Pan J et al. , “Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem,” Eur Radiol Exp, 4(1), 50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tang Y, Gao R, Han S et al. , “Body Part Regression With Self-Supervision,” IEEE Trans Med Imaging, 40(5), 1499–1507 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Modat M, Ridgway GR, Taylor ZA et al. , “Fast free-form deformation using graphics processing units,” Comput Methods Programs Biomed, 98(3), 278–84 (2010). [DOI] [PubMed] [Google Scholar]
- [21].Hamm J, Ye DH, Verma R et al. , “GRAM: A framework for geodesic registration on anatomical manifolds,” Med Image Anal, 14(5), 633–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ying S, Wu G, Wang Q et al. , “Hierarchical unbiased graph shrinkage (HUGS): a novel groupwise registration for large data set,” Neuroimage, 84, 626–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu T, Yang J, Li Q et al. , “Groupwise registration with global-local graph shrinkage in atlas construction,” Med Image Anal, 64, 101711 (2020). [DOI] [PubMed] [Google Scholar]
- [24].Ahlqvist E, Storm P, Karajamaki A et al. , “Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables,” Lancet Diabetes Endocrinol, 6(5), 361–369 (2018). [DOI] [PubMed] [Google Scholar]
- [25].Sobanski V, Giovannelli J, Allanore Y et al. , “Phenotypes Determined by Cluster Analysis and Their Survival in the Prospective European Scleroderma Trials and Research Cohort of Patients With Systemic Sclerosis,” Arthritis Rheumatol, 71(9), 1553–1570 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Oelsner EC, Carr JJ, Enright PL et al. , “Per cent emphysema is associated with respiratory and lung cancer mortality in the general population: a cohort study,” Thorax, 71(7), 624–632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kruskal WH, and Wallis WA, “Use of Ranks in One-Criterion Variance Analysis,” Journal of the American Statistical Association, 47(260), 583–621 (1952). [Google Scholar]
- [28].Rich JT, Neely JG, Paniello RC et al. , “A practical guide to understanding Kaplan-Meier curves,” Otolaryngol Head Neck Surg, 143(3), 331–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cox DR, “Regression Models and Life-Tables,” Journal of the Royal Statistical Society. Series B (Methodological), 34(2), 187–220 (1972). [Google Scholar]
- [30].National Lung Screening Trial Research Team, Aberle DR, Berg CD et al. , “The National Lung Screening Trial: overview and study design,” Radiology, 258(1), 243–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Becht E, McInnes L, Healy J et al. , “Dimensionality reduction for visualizing single-cell data using UMAP,” Nature Biotechnology, 37(1), 38–44 (2019). [DOI] [PubMed] [Google Scholar]


