Abstract
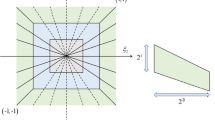
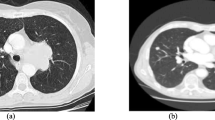
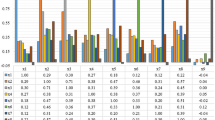
A comprehensive method of B-mode ultrasound image texture analysis for the determination of the liver fibrosis stage is suggested. The algorithm is based on the use of Rotation Forest and KNN classifiers for the texture classification. 720 textural characteristics were extracted using methods based on Laws’ masks analysis, co-occurrence matrix, gray level run-length matrix and statistical characteristics of the images. An optimal subset of 22 informative features was selected using correlation-based method. Testing the algorithm with liver images of 57 patients divided into 5 stages of fibrosis showed 72.7% classification accuracy for single regions of interest. In the case of entire image classification the fibrosis stage was correctly identified for the vast majority of cases.
Similar content being viewed by others
References
Wang, D., Fang, Y., et al., B-scan image feature extraction of fatty liver, in Int. Conf. Internet Comput. Set. Eng. (ICICSE), IEEE, 2012, pp. 188–192.
Virmani, J., Kumar, V., et al., Prediction of cirrhosis from liver ultrasound b-mode images based on laws’ masks analysis, in Int. Conf. Image Inform. Process. (ICIP), IEEE, 2011, pp. 1–5.
Wen-Chun Yeh, Sheng-Wen Huang, and Pai-Chi Li, Liver fibrosis grade classification with b-mode ultrasound, Ultrasound Med. Biol., 2003, vol. 29, no. 9, pp. 1229–1235.
Poonguzhali, S. and Ravindran, G., Automatic classification of focal lesions in ultrasound liver images using combined texture features, J. Inform. Technol., 2008, vol. 7, no. 1, pp. 205–209.
Wen-Li Lee, Yung-Chang Chen, and Kai-Sheng Hsieh, Ultrasonic liver tissues classification by fractal feature vector based on m-band wavelet transform, IEEE Trans. Med. Imaging, 2003, vol. 22, no. 3, pp. 382–392.
Friedrich-Rust, M., Nierhoff, J., Lupsor, M., et al., Performance of acoustic radiation force impulse imaging for the staging of liver fibrosis: a pooled meta-analysis, J. Viral Hepatitis, 2012, vol. 19, no. 2, pp. e212–e219.
Sporea, I., Bota, S., Peck-Radosavljevic, M., et al., Acoustic radiation force impulse elastography for fibrosis evaluation in patients with chronic hepatitis c: an international multicenter study, Eur. J. Radiol., 2012, vol. 81, no. 12, pp. 4112–4118.
Witten, I.H. and Frank, E, Data Mining: Practical Machine Learning Tools and Techniques, 3rd Ed., Morgan Kaufmann, 2011.
Petrou, M. and Sevilla, P.G., Image Processing: Dealing with Texture, Chap. 4.6, Wiley Chichester, 2006.
Haralick, R.M., Statistical and structural approaches to texture, P. IEEE, 1979, vol. 67, no. 5, pp. 786–804.
Selvarajah, S. and Kodituwakku, S.R., Analysis and comparison of texture features for content based image retrieval, Int. J. Latest Trends Comput., 2011, vol. 2, no. 1.
Bouckaert, R. et al., Weka manual for version 3-7-10, 2013.
Cleary, J.G., Trigg, L.E., et al., K*: An instance-based learner using an entropic distance measure, in ICML, 1995, pp. 108–114.
Breiman, L., Random forests, Machine Learning, 2001, vol. 15, no. 1, pp. 5–32.
Rodriguez, J.J., Kuncheva, L.I., and Alonso, C.J., Rotation forest: A new classifier ensemble method, IEEE Trans. Pattern. Anal., 2006, vol. 28, no. 10, pp. 1619–1630.
Wilkinson, L., Anand, A., and Tuan, D.N., Chirp: a new classifier based on composite hypercubes on iterated random projections, in p. 17th ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., ACM, 2011, pp. 6–14.
Melville, P. and Mooney, R.J., Constructing diverse classifier ensembles using artificial training examples, in IJCAI, Citeseer, 2003, vol. 3, pp. 505–510.
Virmani, J., Kumar, V., et al., SVM-based characterization of liver ultrasound images using wavelet packet texture descriptors, J. Digit. Imaging, 2013, vol. 26, no. 3, pp. 530–543.
Balasubramanian, D. et al., Automatic classification of focal lesions in ultrasound liver images using principal component analysis and neural networks, in Ann. Int. Conf. IEEE Eng. Med. Biol Soc., IEEE, 2007, pp. 2134–2137.
Hall, M.A., Correlation-based feature selection for machine learning, PhD Thesis, The University of Waikato, 1999.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Kvostikov, A.V., Krylov, A.S. & Kamalov, U.R. Ultrasound image texture analysis for liver fibrosis stage diagnostics. Program Comput Soft 41, 273–278 (2015). https://doi.org/10.1134/S0361768815050059
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0361768815050059